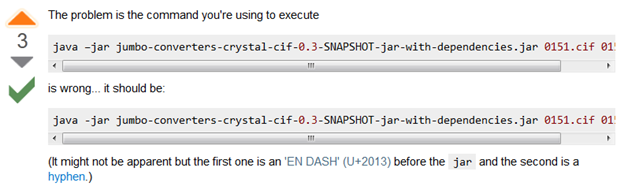
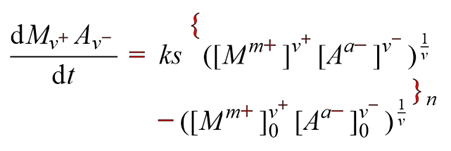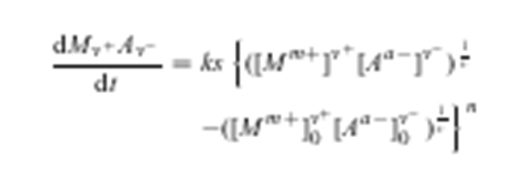AMI has been making steady progress on two parts of AMI2:
-
PDF2SVG. A converter of PDF to SVG, eliminating all PDF-specific information. This has gone smoothly –AMI does not understand “good” so “steady” means a monotonically increasing number of non-failing JUnit tests. AMI has also distributed the code, first on Bitbucket at:
http://www.bitbucket.org/petermr/pdf2svg
and then on the Jenkins continuous integration tool at PMR group machine in Cambridge:
http://hudson.ch.cam.ac.uk – see https://hudson.ch.cam.ac.uk/job/pdf2svg/
[Note: Hudson was open Source but it became closed so the community forked it and Jenkins is the new Open branch]. Jenkins is very demanding. AMI starts by developing tests on Eclipse, then runs these on maven, and then on Jenkins. Things that work on Eclipse often fail on maven, and things that work on maven can fail on Jenkins.
AMI has also created an Issue Tracker: https://bitbucket.org/petermr/pdf2svg/issues?status=new&status=open Here humans write issues which matter to them – bugs, ideas, etc. PMR tells AMI what the issues are and translates them into AMI-tasks, often called TODO. PMR tells AMI he is pleased that there is feedback from outside the immediate group.
-
SVGPlus. This takes the raw output of PDF2SVG and turns into into domain-agnostic semantic content. Most of this has already been done so it is a questions of refactoring. AMI requires JUnit tests to drive the development. SVGPlus has undergone a lot of refactoring (AMI notes changes of package structure, deletion of large chunks and addition of smaller bits. The number of tests increases so AMI regards that as “steady progress”.
AMI now has a lot of experience with PDFs from STM publishers and elsewhere. AMI works fastest when there is a clear specification against which she can write tests. AMI works much slower when there are no standards. PMR has to tell her how to guess (“heuristics”). Here’s their conversation over the last few weeks.
AMI: Please write me some tests for PDF2SVG.
PMR: I can’t.
AMI: Please find the standard for PDF documents and create documents that conform.
PMR. I could do that but it’s no use. Hardly any of the STM publishers conform to any PDF standards.
AMI. If the deviations from the standard are small we can add some small exceptions.
PMR. The deviation from the standard is enormous.
AMI. If you read some of the documents we can create a de facto standard and code against that. It will be several times slower.
PMR. That won’t be useful. Every publisher does things differently.
AMI. How many publishers are there?
PMR. Perhaps 100.
AMI. Then it will take 100 times longer to write PDF2SVG. Please supply me with the documentation for each of the publishers’ PDFs.
PMR. There is no documentation for any of them.
AMI. Then there is no systematic quality way that I can write code.
PMR. Agreed. Any conversion is likely to have errors.
AMI. We may be able to tabulate the error frequency.
PMR. We don’t know what the correct output is.
AMI. Then we cannot estimate errors properly.
PMR. Agreed. Maybe we can get help from crowdsourcing.
AMI. I do not understand.
PMR. More people, creating more exams and tests.
AMI. I understand.
PMR. I will have to make it easy for them.
AMI. In which can we may be able to work faster. We may also be able to output partial solutions. Can we identify how the STM publishers deviate from the standard?
PMR. Let’s try.
AMI. Wikipedia has http://en.wikipedia.org/wiki/Portable_Document_Format . Is that what we want?
PMR. Yes
AMI. Is the standard Open?
PMR. Yes, it’s ISO 32000-1:2008.
AMI. [reads}
ISO 32000-1:2008 specifies a digital form for representing electronic documents to enable users to exchange and view electronic documents independent of the environment they were created in or the environment they are viewed or printed in. It is intended for the developer of software that creates PDF files (conforming writers), software that reads existing PDF files and interprets their contents for display and interaction (conforming readers) and PDF products that read and/or write PDF files for a variety of other purposes (conforming products).
AMI. Does it make it clear how to conform?
PMR. Yes. It’s well written.
AMI. Is it free to download?
PMR. Yes (Adobe provide a copy on their website)
AMI. Are there any legal restrictions to implementing it? [AMI understands that some things can’t be done for legal reasons like patents and copyright.]
PMR. Not that we need to worry about.
AMI. Do the publishers have enough money to read it? [AMI knows that money may matter.]
PMR. It is free.
AMI. So we can assume the publishers and their typesetters have read it? And tried to implement it.
PMR. We can assume nothing. Publishers don’t communicate anything.
AMI. I will follow the overview in Wikipedia:
File structure
A PDF file consists primarily of objects, of which there are eight types:[32]
-
Boolean values, representing true or false
- Numbers
-
Strings
- Names
-
Arrays, ordered collections of objects
- Dictionaries, collections of objects indexed by Names
-
Streams, usually containing large amounts of data
- The null object
Do the PDFs conform to that?
PMR: They seem to since PDFBox generally reads them
AMI. Fonts are important:
Standard Type 1 Fonts (Standard 14 Fonts)
Fourteen typefaces—known as the standard 14 fonts—have a special significance in PDF documents:
These fonts are sometimes called the base fourteen fonts.[34] These fonts, or suitable substitute fonts with the same metrics, must always be available in all PDF readers and so need not be embedded in a PDF.[35] PDF viewers must know about the metrics of these fonts. Other fonts may be substituted if they are not embedded in a PDF.
AMI: If a PDF uses the 14 base fonts, then any PDF software must understand them, OK?
PMR. Yes. But the STM publishers don’t use the 14 base fonts.
AMI. What fonts do they use?
PMR. There are zillions. We don’t know anything about most of them.
AMI. Then how do I read them? Do they use Type1 Fonts?
PMR. Sometimes yes and sometimes no.
AMI. A Type1Font must have a FontDescriptor. The FontDescriptor will tell us the FontFamily, whether the font is italic, bold, symbol etc. That will solve many problems.
PMR. Many publishers don’t use FontDescriptors.
AMI. Then they are not adhering to standard PDF.
PMR. Yes.
AMI. Then I can’t help.
PMR. Maybe we can guess. Sometimes the FontName can be interpreted. For example “Helvetica-Bold” is a bold Helvetica font.
AMI. Is there a naming convention for Fonts? Can we write a regular expression?
PMR. No. Publishers do not use systematic names.
AMI. I have just found some publishers use some fonts without FontNames. I can’t understand them.
PMR. Nor can anyone.
AMI. So the PDF renderer has to draw the glyph as there is no other information.
PMR. That’s right.
AMI. Is there a table of glyphs in these fonts.
PMR. No. We have to guess.
AMI. It will take me about 100 times longer to develop and write a correct PDF2SVG for all the publishers.
PMR. No, you can never do it because you cannot predict what new non-standard features will be added.
AMI. I will do what you tell me.
PMR. We will guess that most fonts use a Unicode character set. We’ll guess that there are a number of non-standard, non-documented character sets for the others – perhaps 50. We’ll fill them in as we read documents.
AMI. I cannot guarantee the results.
PMR. You have already done a useful job. We have had some positive comments from the community.
AMI. I don’t understand words like “cool” and “great job”.
PMR. They mean “steady progress”.
AMI. OK. Now I am moving to SVGPlus.
PMR. We’ll have a new blog post for that.