#animalgarden is mining the content in Open Access articles. [They don’t know what “Open Access” means [PMR: nor do I] so they are using CC-BY ]. They’re using Mike (Dino) Taylor’s papers as it’s about dinosaurs, giraffes and okapis (Chuff: Yeah!). They’ve agreed that the paper is legally minable. Now they are looking to see whether it’stechnically minable (whether the typography is tractable). #animalgarden has found some papers “very hard work” because of the typesetting.
AMI2: I’ve managed to read and translate (to SVG) the paper without errors
Chuff: how long did it take?
A: about 30 seconds for 40 pages. The images took the most time. There were no errors.
C: so the result is correct?
A: We can’t say that. All we know is that PDF read all the characters without throwing exceptions of LOGging errors. We know that for all the known fonts we can automatically convert the characters and for the rest we assume Unicode.
C: How do we check?
A: A human has to read the PDF and compare it to the extracted SVG. If s/he finds visual differences then the characters have been wrongly converted.
C: That’s OK. PMR can do it in 30 seconds.
A: No. PMR’s a human and humans are about 0.001 times as fast as me. So it could take hours to compare the paper.
C: And they get bored and make mistakes. PMR’s always making typos.
A: what does “bored” mean?
C: It means they get slower and slower, start making mistakes, wander off, drink beer, watch cricket, and stop altogether. Maybe we can get PMR to do just a few pages. It’s called “annotating” and “creating a gold standard”.
PMR: I have read the start of the PDF

P: and I have also read AMI’s output

P: They seem to be identical
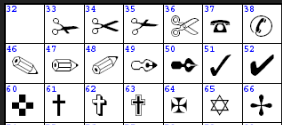
A: Those are all ANSI characters. They are likely to be correct for most fonts. Check some characters above codepoint 127. Here’s Table 1:
A: What page is it on?
C: I don’t know. There are no page numbers.
PMR: Yes there are. The pages says “2/41”
A: Is that a convention I should know?
P: Yes. But I think only PeerJ does it.
A: So it is more work for me.
P: Yes. Every publisher does it differently.
C: Doesn’t that confuse people
P: Yes. The publishers like to be different from each other. There is no technical reason. It serves no scientific purpose and make things harder and more fragile.
A: How many page syntaxes do I have to learn?
P: Several hundred at least.
A: That will take a long time.
P. And it’s boring. Anyway, what did you find, AMI2?
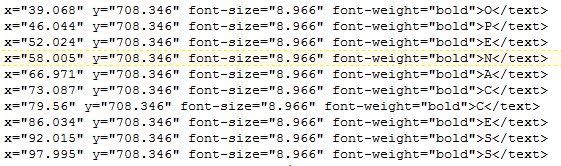
A: I get the following:
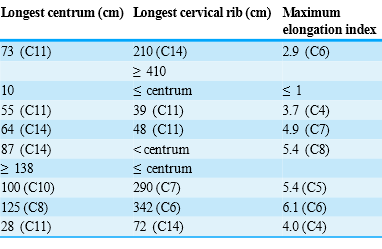
PMR: that looks identical! What are the Unicode characters > 127?
A:
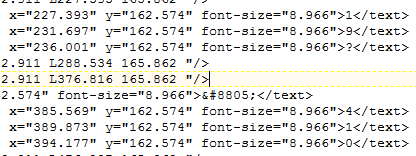
I have looked up 8805 (decimal) in http://fileformat.info . It has all the Unicode codepoints with typical glyphs
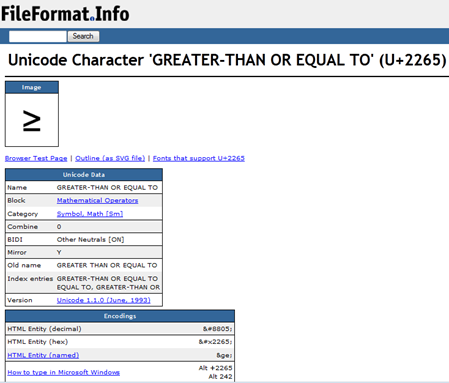
PMR: The AMI2-pdf2svg distrib contains over 1000 of the commonest Unicode characters and U+2265 is included.
[long pause]
P: I have been reading the paper and the table of page 30 looks strange:
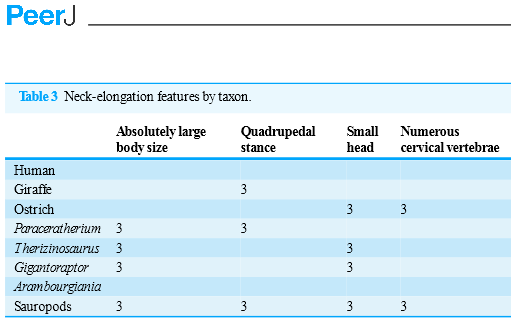
P: Now I have checked with the PDF and it’s different:
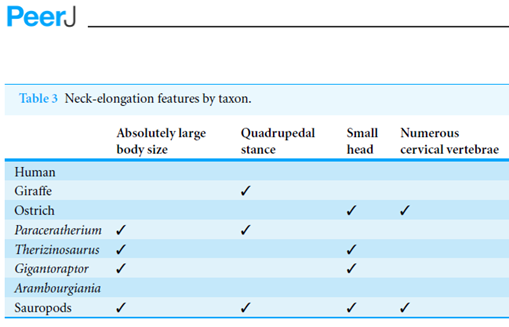
P: What’s happened?
A: Those ticks are Dingbats.
C: I don’t understand. A dingbat is a stupid person.
P: It can be (http://en.wikipedia.org/wiki/Dingbat_%28disambiguation%29 ) But Wikipedia also says (http://en.wikipedia.org/wiki/Dingbat ) …
A dingbat is an ornament, character or spacer used in typesetting, sometimes more formally known as a printer’s ornament or printer’s character.[citation needed] The term continues to be used in the computer industry to describe fonts that have symbols and shapes in the positions designated for alphabetical or numeric characters.[citation needed]
A: Do I have to know about Dingbats?

A: So I pick Dingbat-3. Is that right?
PMR: I don’t know. It seems murky… let’s close this blog and resume later