PMR has been hacking bugs with MJ and AMI2…
PMR: PDF2SVG should now manage Type0 fonts and we’ve fixed bugs on some character processing. Now let’s look at the Dingbats… AMI, do you have a Dingbat lookup-table?
A: No
P: OK, we’ll have to create one. I’ve tried to find a conversion table to Unicode on the web… but failed. However we’ve found:
(that’s only part because the whole picture might be copyright).
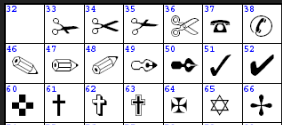
A: so what’s the Unicode for (char)51?
P: It’s a “tick”. I’ll have to look it up in the Unicode
A: I can’t understand glyphs yet. You’ll have to do it for me. There’s about 170. Is that “boring”?
P: VERY. But I will only do the ones I need and hope others will help out. It only has to be done once. I’ve found:

They don’t match up so I’ll have to do them one-by-one… AMI, please would you create a new table “dingbats.xml” in
pdf2svg1/src/main/resources/org/xmlcml/pdf2svg/codepoints/misc/dingbats.xml
A: done.
P: we’ll map char-51 to U+2713 because they seem to be the same (and char-52 to U+2714, etc.. And while I’m watching the cricket I can do some more. We have now g confidence in converting 99.99+% of the characters that are likely to occur in bioscience. There’s more work to do for maths.
A: Yes. I have a document which fails with CambriaMath. We default to Unicode so CambriaMath-4666 defaults to ETHIOPIC SYLLABLE SHI (U+123A) which looks like:

Is that what you want?
P: No I have to translate each character into the table
A: There are only 10000 so that should take a few milliseconds…
P: SHI
this might help – maybe
ftp://ftp.unicode.org/Public/MAPPINGS/VENDORS/ADOBE/zdingbat.txt