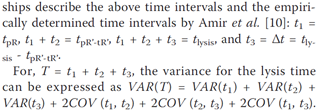On Saturday we are having an economics hackathon in London. I’d love to be there but unfortunately am going to the Eur Sem Web Conf in Montpelier. It’s run by Velichka and colleagues – here’s the sort of reason why (from OKFN blog)
Velichka Dimitrova revisted the disgraced Reinhart-Rogoff paper on austerity economics, the perfect evidence of the need for open data in economics – and was picked up by the London School of Economics and the New Scientist.
The point is that economists made very serious mistakes and that proper management of the data and tools could have prevented it. We have to work towards reproducible computation in sciences and economics. From Velichka’s blog (and then I set you a puzzle at the end):
Another economics scandal made the news last week. Harvard Kennedy School professor Carmen Reinhart and Harvard University professor Kenneth Rogoff argued in their 2010 NBER paper that economic growth slows down when the debt/GDP ratio exceeds the threshold of 90 percent of GDP. These results were also published in one of the most prestigious economics journals – the American Economic Review (AER) – and had a powerful resonance in a period of serious economic and public policy turmoil when governments around the world slashed spending in order to decrease the public deficit and stimulate economic growth.
Yet, they were proven wrong. Thomas Herndon, Michael Ash and Robert Pollin from the University of Massachusetts (UMass) tried to replicate the results of Reinhart and Rogoff and criticised them on the basis of three reasons:
- Coding errors: due to a spreadsheet error five countries were excluded completely from the sample resulting in significant error of the average real GDP growth and the debt/GDP ratio in several categories
- Selective exclusion of available data and data gaps: Reinhart and Rogoff exclude Australia (1946-1950), New Zealand (1946-1949) and Canada (1946-1950). This exclusion is alone responsible for a significant reduction of the estimated real GDP growth in the highest public debt/GDP category
- Unconventional weighting of summary statistics: the authors do not discuss their decision to weight equally by country rather than by country-year, which could be arbitrary and ignores the issue of serial correlation.
The implications of these results are that countries with high levels of public debt experience only “modestly diminished” average GDP growth rates and as the UMass authors show there is a wide range of GDP growth performances at every level of public debt among the twenty advanced economies in the survey of Reinhart and Rogoff. Even if the negative trend is still visible in the results of the UMass researchers, the data fits the trend very poorly: “low debt and poor growth, and high debt and strong growth, are both reasonably common outcomes.”
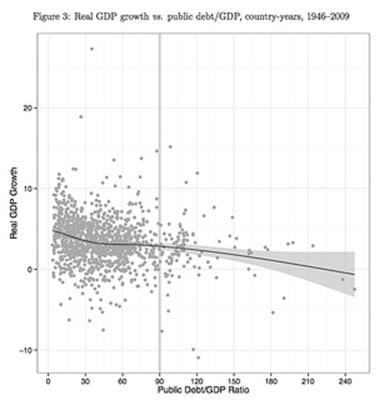
Source: Herndon, T., Ash, M. & Pollin, R., “Does High Public Debt Consistently Stifle Economic Growth? A Critique of Reinhart and Rogoff, Public Economy Research Institute at University of Massachusetts: Amherst Working Paper Series. April 2013.
What makes it even more compelling news is that it is all a tale from the state of Massachusetts: distinguished Harvard professors (#1 university in the US) challenged by empiricists from the less known UMAss (#97 university in the US). Then despite the excellent AER data availability policy – which acts as a role model for other journals in economics – the AER has failed to enforce it and make the data and code of Reinhart and Rogoff available to other researchers.
Coding errors happen, yet the greater research misconduct was not allowing other researchers to review and replicate the results through making the data openly available. If the data and code were made available upon publication in 2010, it may not have taken three years to prove these results wrong, which may have influenced the direction of public policy around the world towards stricter austerity measures. Sharing research data means a possibility to replicate and discuss, enabling the scrutiny of research findings as well as improvement and validation of research methods through more scientific enquiry and debate.
So Saturday’s hackathon (I might manage to connect in on Eurostar?) is about building reliable semantic models for reporting economics analyses. Since economics is about numbers and chemistry is about numbers there’s a lot in common and the tools we’ve developed for Chemical Markup Language might have some re-usability. So this morning Velichka, Ross Mounce and I had a skype to look at some papers.
We actually spent most of the time on one:

And here’s one example of a data set in the paper. (Note it’s behind a paywall (JSTOR) and I haven’t asked permission and I don’t need to tell you about what happened between Aaron Swartz and JSTOR. But I argue that these are facts and fair cricitism):
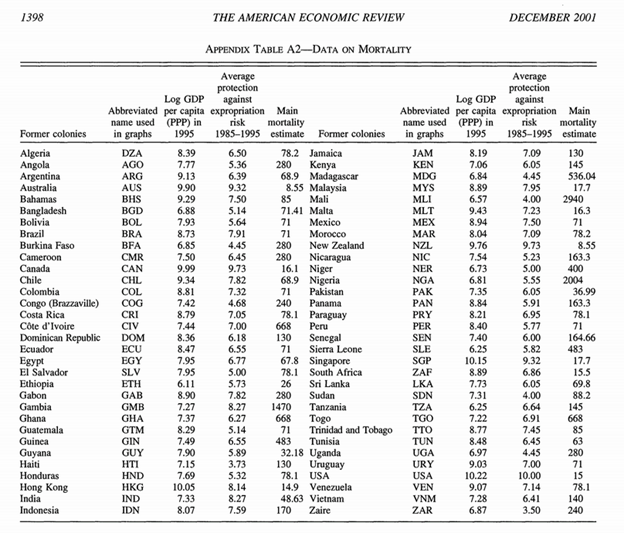
The authors regressed the dependent variable (log GDP) against the other two. My questions, as a physical scientist are:
- What are the units of GDP? After all someone might use different ones. And I personally cannot understand the values.
- What is “main mortality estimate”? If you guess without reading the paper you will almost certainly be wrong. You have to read the paper very carefully and even then take a lot on trust.
I’m not suggesting that this research is reproducible just from this table (although it should be possible to regenerate the same results). I’m arguing that data of this sort (I exclude it as 12 years old) is not acceptable any more. Data must be unambiguously labelled with units and described so the source and data are replicable.