Two impressive blog posts from Cameron Neylon on Open Notebook Science:
So I have given three talks in ten days or so, one at the CanSAS meeting at NIST, one at Drexel University and one at MIT last night. Jean-Claude Bradley was kind enough to help me record the talk at Drexel as a screencast and you can see this in various formats here. He has also made some comments on the talk on the UsefulChem Blog and Scientific Blogging site.
The talks at Drexel and MIT were interesting. I was expecting the focus of questions to be more on the issues of being open, the risks and benefits, and problems. Actually the focus of questions was on the technicalities and in particular people wanting to get under the hood and play with the underlying data. Several of the questions I was asked could be translated as ‘do you have an API?’. The answer to this is at the moment no, but we know it is a direction we need to go in.
We have two crucial things we need to address at the moment: the first is the issue of automating some of the posting. We believe this needs to be achieved through an application or script that sits outside the blog itself and that it can be linked to the process of actually labelling the stuff we make. The second issue is that of an API or web service that allows people to get at the underlying data in an automated fashion. This will be useful for us as we move towards doing analysis of our data as well. Jean-Claude said he was also looking at how to automate processes so clearly this is the next big step forward.
Another question raised at MIT was how you could retro-fit our approach into an existing blog or wiki engine. The key issues here are templates (which is next on my list to describe here in detail) which would probably require some sort of plugin. The other issue is the metadata. Our blog engine goes one step beyond tagging by providing keys with values. Presumably this could be coded into a conventional engine using RDF or microformats – perhaps we should be doing this our Blog in any case?
Incidentally a point I made in both talks, partly in response to the question ‘does anyone really look at it’, is that in many cases it is your own access you are enabling. Making it open means you can always get at your own data, which is a surprisingly helpful thing.[…]
[…]
- Neither Wikis nor Blogs provide all the functionality required. Wikis are good at providing a framework that within which to organise information where as blogs are good at logging information and providing it in a journal format. Barry showed me a hack that he uses in his Wiki based notebook that essentially provides a means of organising his lab book into experiments and projects but also provides a date style view. In the Southampton system we would achieve this through creating categories for different experiments, possibly independent blogs for different projects.
- Feature requests at Southampton has been driven largely by me which means that system is being driven by the needs of the PI. At OpenWetWare the development has been driven by grad students which means it has focussed on their issues. The question was raised of where the best place to ‘promote’ these systems was. Is it the PI’s who, at least at the moment, will get the greatest tangible benefits from the system. Or is it better to persuade grad students to take this up as they are the end users. Both have very different needs.
- Development based on the needs of a single person is unlikely to take us forward as the needs of a specific person are probably not general enough to be useful. Development should focus on enabling the interactions between people, therefore the minimum size ‘user unit’ is two (PI plus researcher, or group of researchers).
- The biggest wins for these systems are where collaboration is required and is enabled by a shared space to work in. This is shown by the IGEM lab books and by uptake by my collaborators in the UK. This will be the best place to take development forward.
I need to add links to this post but will do so later. [PMR – know the feeling…]
I agree with the idea that the primary beneficiary is often the author or the team, rather than the world. I also realise how much implicit information there is – it’s very difficult to make everything available instantly. For example in the computational area we should theoretically expose evidence of submitting jobs but we often don’t keep this for ourselves, relying on the system to manage it for us. It brings home how much work we need to do to document fully what we do.
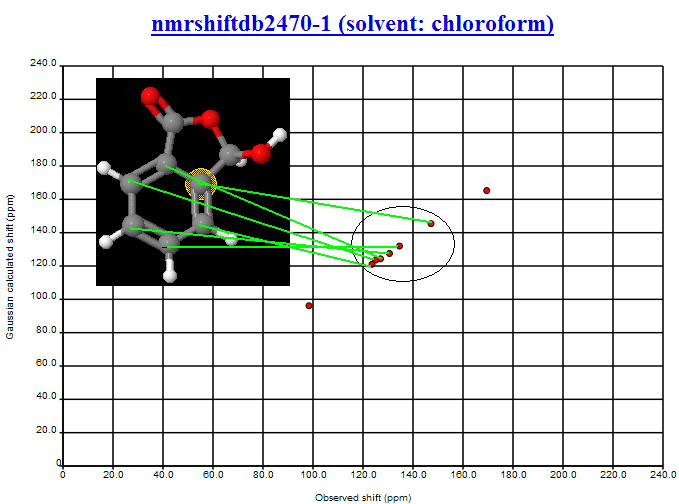
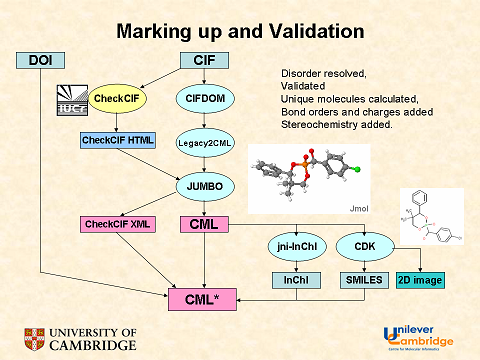
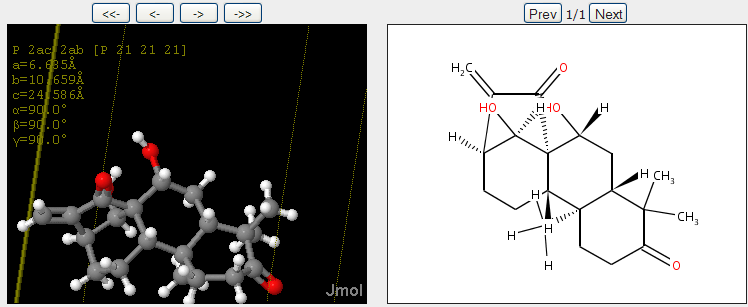
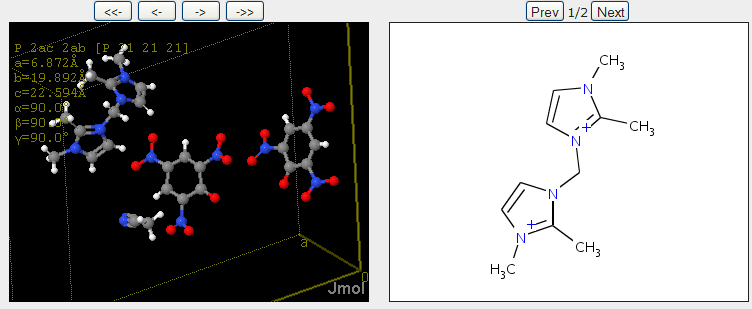
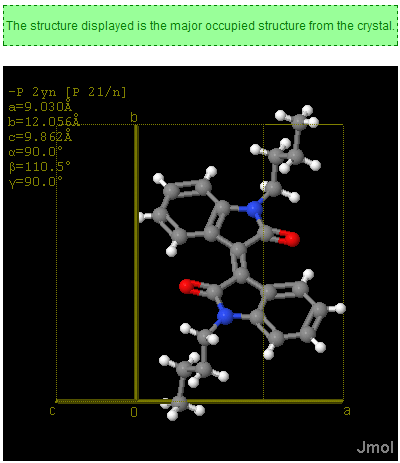
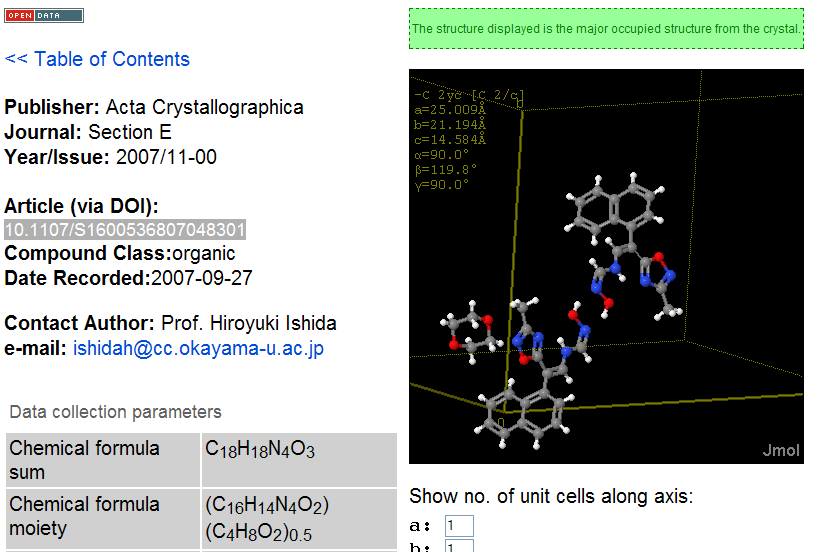

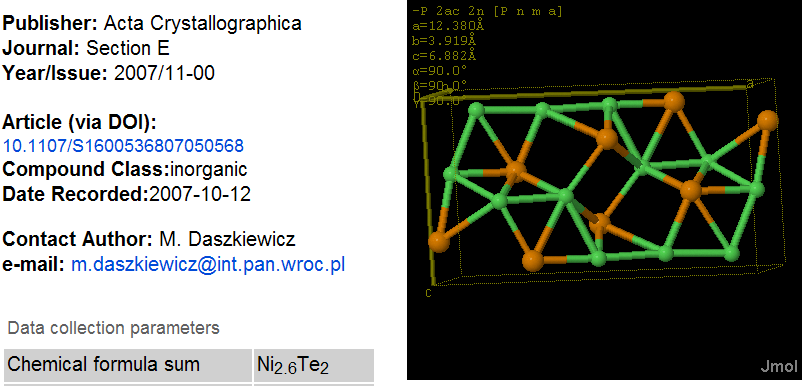
November 4th, 2007 at 1:43 pm eRegarding the InChIs: I would prefer one InChI for each moiety, not one InChI for the full structure. Or not only, at least.