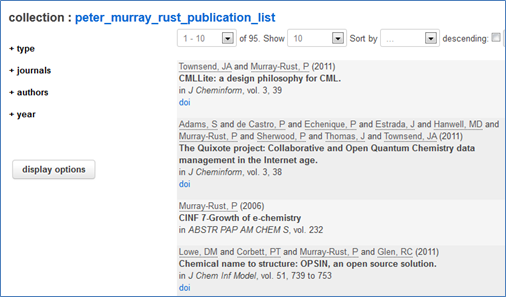
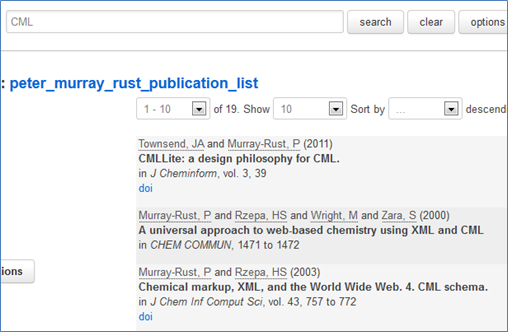
#pantonfellowships #pantonprinciples

In 2010 we launched the Principles of Open Scientific Data and, because we met more than once in the Panton Arms we called them the “Panton Principles”. Since then “Panton” has started to become a brand for Openness. We’ve now had 6 Panton discussions with people who champion Open Data, and we are creating Panton Papers to support the discussion and formulation of ideas in Open Data.
Now we take this a major step forward. The idea to create Fellowships came from Jonathan Gray and he and I worked up an application to the Open Society Foundations (previously called OSI http://en.wikipedia.org/wiki/Open_Society_Institute ) . We were delighted that in November the OSF told us that the grant had been successful, and we thank them.
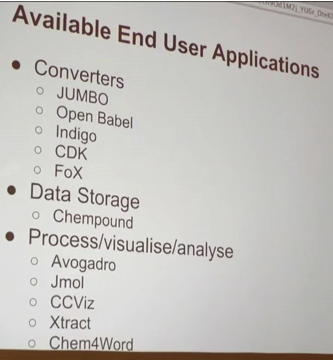
The idea of the fellowships is to support scientists to develop activities in Open Data. The remit is deliberately broad as we want to encourage novel approaches. The details are given here. http://blog.okfn.org/2012/01/25/panton-fellowships-apply-now/ with an excerpt:
We firmly believe that “open data means better science“. Panton Fellowships have been created in order to support scientists – particularly graduate students and early-stage career scientists – to explore this idea, and to tackle those barriers which currently prevent science data from being made open.
Dr Cameron Neylon, of the Panton Fellowships Advisory Board, commented on the ‘real potential’ of the Fellowships to influence practice surrounding open data in the scientific community. ‘Panton Fellowships will allow those who are still deeply involved in research to think closely about the policy and technical issues surrounding open data’, observed Dr Neylon. By allowing scientists the scope both to explore the ‘big picture’ – gathering evidence to promote discussion throughout the community – and also to work on specific technical solutions to individual problems, the Panton Fellowship scheme has the potential to make a real impact upon the practice of open data in science.
Panton Fellows will have the freedom to undertake a range of activities, and prospective applicants are encouraged to formulate their own work plan. As Fellows will continue to be employed and/or study at their current institution, activities undertaken for the Panton Fellowship should ideally complement and enhance their existing work.
Fellowships will be held for one year, and will have a value of £8k p.a. For more details and information on how to apply, please visit http://pantonprinciples.org/panton-fellowships/.
The Panton philosophy is increasingly adopted by promoters of Openness and as an example BiomedCentral have donated us banner advertising on their pages.
I am really looking forward to reading the applications!