I have worked with computer graphics for many years – this has been catalysed by Noel’s discussion of the Algorithm puzzles
This brings back lots of memories. I remember reading a strange report in (I think) nature about a mysterious function (later to be called the Mandelbrot set). It’s very simple – takes about 5 lines of code. So I set it up on the BBC. (For non-UK readers, in the eighties a wonderful home and educational computer, with an excellent mix of operating system, assembler, graphics and Basic interpreter (though “Basic” doesn’t do it credit).
Anyway I saw this function and wrote a little program to iterate. I ran it and couldn’t see anything special. So I thought I needed a finer grid. It was going to take about 12 hours to draw the picture (not the drawing speed – the iterations through the complex numbers). I didn’t expect but left it on overnight. I came in in the morning, having forgotten that I’d left it on, and got one of the wow! factors of my life:
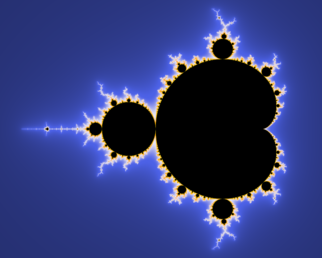 (Thanks, Wikipedia)
(Thanks, Wikipedia)
I think mine was in monochrome, but still wow!
The great thing about the Beeb was that you could just sit down and run the graphics. That was also true on the AppleII (my first personal machine – well actually the lab’s but all mine!). Now you can’t. If I want to draw some graphics what do I do?
For a quick lash up I might generate an SVG and then view it – but that’s a bit clunky. But otherwise? How do I type move and draw on the commandline? It’s got worse.
I’ve been through the whole range of graphics software – Tek4010, Calcomp plotter, AppleII, Megatek, Evans and Sutherland PS300, BBC, VT200, SGI-GL, GKS, PHIGS, OpenGL, TK, Java AWT1.02, Swing, Java3D, Swing… and some I’ve happily forgotten. If I want reasonable high-quality interactive graphics I suppose I’d choose Swing. I hate Swing. Typical error message “You cannot add a JPanel to JFrame, you must add to a JContentPane” – absurd! The logic is clear only to the authors (if anyone). If some meaningless intermediate object is required, why can’t the software add it? And you have to remember when to use pack() and show() and …The one I remember with most fondness was TK – well ahead of its time.
So I can’t answer Noel’s problem in 5 minutes – it’s half a day or downloading libraries and working out how to use them. Or not. SVG is great, but I can’t just play with it. And I know that I and my colleagues need to start using some graphics. Where should we start…


November 30th, 2006 at 1:59 pm eI would be *very* impressed if you could figure out the solution without running the program. Try coding it – a picture tells a thousand words.
November 30th, 2006 at 2:55 pm e(3) All right. I have to decide yet again what graphics system I should aim for – GTK, Swing, or even SVG.
My latest guess is a Cantor set of some sort (perhaps a Sierpinski gasket) with an empty triangle in the middle
November 30th, 2006 at 5:44 pm eIn the old days, this would take 5 minutes on a BBC to code up and run. Even in Windows with Qbasic, it was easy. The barrier is higher now…Python requires an additional module such as PyGame.