Data in chemistry publications is very standardized which makes it possible (not easy) to think about robotic extraction of information. I’ve blogged earlier about the use of text, but what about graphics? This post shows the potential, but also the current unnecessary destruction of data. You don’t need to be a chemist to understand the issue.
Types of graphical object that occur frequently in chemistry are
- chemical structure diagrams (more later)
- graphs (i.e. plots, not topology, though these also occur)
- spectra (used to probe the nature of compounds and also to act as fingerprints.
Here I show some proton-NMR spectra (1H NMR), which are very powerful ways of looking into molecules containing hydrogen atoms (almost all do). It’s closely related to NMRI used for medical imaging. What is remarkable is its precision – the frequency used is (often) 500 Mhz (i.e. 5 * 108 per second. Because of this precision the frequency axis is usually expressed in parts per million (ppm). The scale runs from 10 to 0 ppm. This is recorded digitally, usually with 2N points, such as 8192, 16384 or even more. So that means that for each ppm there are about 1000 points or more.
The values and the precise shapes of the peaks are very important. They are usually quoted to 2 decimal places and the fine structure (“coupling”) can be meaningful even if as small as 1 Hz (i.e. 0.02 ppm).
In the SPECTRa-We’ve been looking at how we can preserve this valuable data – it comes out of the machine in digital form, but then it is often transcribed into a PDF. Sometimes this preserves the graphics structure, sometimes it converts it to a pixellated image. This is the worst sort of hamburger.
Since the spectra are important tools in ensuring reproducibility, and chemists frequently refer to literature values, why do some journals allow such awful spectra. I suppose it’s better than having no spectra at all. Here are some good bad and ugly from supplemental info for recent synthetic chemistry papers. Since at least 3 of them carry a copyright I shan’t identify the journals. I claim that they are (a) data (b) a small portion of the work (c) publication does not affect sales (d) that most people would be ashamed to copyright them anyway.
Note that they all cover about 1 ppm (although for some you have to take the numerals on trust)
Fig. 1 The fuzz is real, but quite a bit is visible
Fig.2 Good. this seems to have preserved most of the data
Fig. 3 What are those figures??? Yes, I can guess – but I shouldn’t have to. But the limited pixel resolution has destroyed the peak shapes as well. Look at the non-linearity of the horizontal axis.
Fig 4. I’ve made this larger so the fuzziness from the pixellation is revealed.
Fig. 5 Quite good. You can certainly see peaks separated by 1 Hz.
Fig 6. Oh dear. This has the added fun of being a JPG which adds some dots to the spectrum which are nothing to do with the data. JPGs should not be used for this sort of thing.
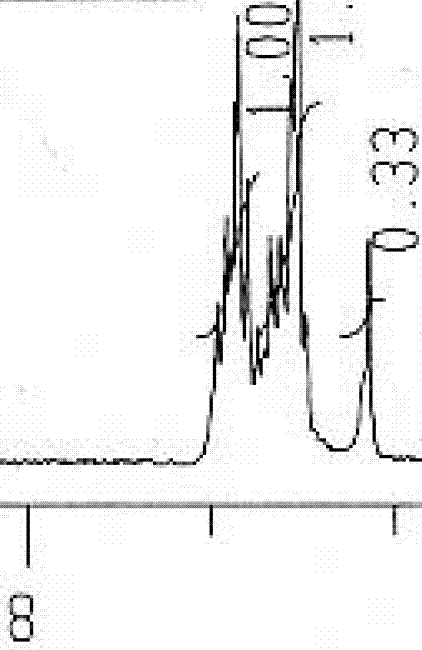
Fig 7. This is 8-7 ppm. Another JPG
So non-chemists should be able to see the point. If an article costs USD 3000 then the scientific community deserves better. How many chemists have cursed the unreadability of numeric data mangled by graphics tools? There is no technical reason why the digital data shouldn’t be deposited with the publisher, the instituion, the department.
The simple question is: do chemists care?






![[DSC00450]](http://farm2.static.flickr.com/1131/1032745399_249144e8ac.jpg)




August 7th, 2007 at 1:23 “In their previous papers, they had to use a metal to take diphosgene to dichloroketene, but in this case, a bit of ultrasound worked rather well.”Ultra sound avoids the use of activated zinc (ref 14), but you definitely need a metal.
Maybe I am wrong but there is no supp info available, as often with Angewandte
August 7th, 2007 at 1:58 spiro, the supporting info is available at http://www.wiley-vch.de/contents/jc_2002/2007/z702031_s.pdfand i think the procedure for the 2+2 is found in their previous methodology paper,referenced in this one.
August 7th, 2007 at 2:49 aa, thanks for your dedication, but I had read these “supporting information” before writing my discontentment.
It is just that I do not consider this to be a decent supporting information section, even though the three procedures they show are the most important of the article.
I do not blame the authors, just the journal. If my boss tells me to write a paper without supp info, I cheer. But this is a bad habit IMHO.
For example, I am perplex about transformation c in scheme 3, especially when I read ref 19. One way or another there may be something which is missing in the conditions (acid?), and a written procedure could clarify things.
August 7th, 2007 at 3:09 The lack of supp info in ACIEE is really frustrating. If you’ve done a total synthesis, why can’t the supp info include any procedures for making compounds not already found in the literature? If I’m doing a lit search, and I find a reaction in Org Lett that I can use, I cheer because it will have a procedure most likely. If it’s for ACIEE, I groan, because the supp infos are so spotty.
August 7th, 2007 at 14:10 I totally agree with you carbazole, I cannot conceive that in 2007, a supp. info for a total synthesis includes only 2 procedures and 4 nmr. It is clearly a lack of rigour from Angew…
August 7th, 2007 at 15:31 spiro- right, sorry about that. yes, the lack of SI in ACIE is pretty terrible. i especially hate when their a reaction in a tot syn that you would like to try and can’t get a detailed procedure.
August 7th, 2007 at 15:52 Supporting Info is all that really matters in these papers anyway right? I mean its nice that someone made something, but its irrelevant if irreproducible bc of spotty SI.
August 7th, 2007 at 16:15 [previous]: The ultimate best example was last year’s synthesis by James La Clair… Deoxoudol, or the molecule-that-shall-change-name-upon-criticism-of-its-synthesis! [1]
August 7th, 2007 at 16:42 Isn’t an author obliged to provide experimental detail/spectral data on request for published reactions? My ex-boss certainly behaves like it – and believe me, the hours I put into scanning spectra and compiling supp. info means the SI isn’t “spotty” in the slightest.
August 8th, 2007 at 4:40 Sure, being required to provide spectra/procedures on request is fine, but why can’t it be included online at the time of publication? Are their servers running low on hard disk space? It was different before online publication obviously, journal pages were precious. Why should I have to email someone for something that really should be provided in the first place?
August 8th, 2007 at 5:31 It makes me wonder if that might be why so often high level papers get sent to Angew over JACS by certain groups in particular….
August 8th, 2007 at 15:56 Jose is onto something.Anyone see baran’s SI for Chartelline in JACS. It was immaculate, the way SI should be.
August 8th, 2007 at 18:23 Most of the time when I email someone for supporting info I don’t get it.. I emailed one of sharpless’s underlings for SI on allyic azide precursors and got not a single response.