Things are happening so fast on the Open Notebook NMR project that we need to take stock. Here’s today’s developments:
- Nick Day is working round the clock to manage the data and create the plots. He has a thesis to write 🙂
- Henry Rzepa continues to have near-daily insights into the NMR methodology
- Antony Williams and now ACD labs would like to work with the data
- Christoph Steinbeck (NMRShiftDB) is visiting today
This highlights the problem of the process of discovery. The most important person is Nick. He has spent the last year developing this software (much of it re-used from CrystalEye) and now he’s got a chance to use it. It’s similar to instrument-based physics where a student spends three years developing a new type of detector and has a mad rush to point it at the sky and show it works. Then it’s released to the world and they start to make their own independent discoveries. So we have to have a delay where Nick can discover things. That’s one reason why the two of us sat down yesterday and published all the things we might expect to discover. Then, when we or others find them, Nick can claim all or some of the credit.
Similarly Henry has developed a number of new insights into NMR. They are his, and if I published them here without his agreement it would be inappropriate. So at this stage I will simple say that Henry has an important protocol that explains a large percentage of the variance for certain chemical groups and I have asked him to publish it here or send the material.
Christoph certainly has the right to have a few day’s breathing space while we look at what’s in NMRShiftDB. He and colleagues have spent years building it up and if we have a new tool then they should get the chance to use it on their data.
Antony and colleagues have offered help and this is welcomed.
We hadn’t anticipated this and we have to think about how this fits in. This is a structured project, on which parts of Nick’s thesis are based, and if other people do some of the work he had planned it causes problems. I think we can work it out but we need to set the ground rules publicly.
There is also the role of ACDLabs which I consider in a later post.
So over the next week we shall probably share our insights but not our data. Probably the first thing we shall ask the community for is help with outliers – here is a structure which doesn’t fit – does anyone know why? Henry has already done this for the worst outlier and shown that it is probably a transcriptional or related error – i.e. there is a report of the compound in the literature which has relatively unremarkable values. If, during this process or weeding out outliers someone discovers a completely new effect then they will deserve a lot of credit.
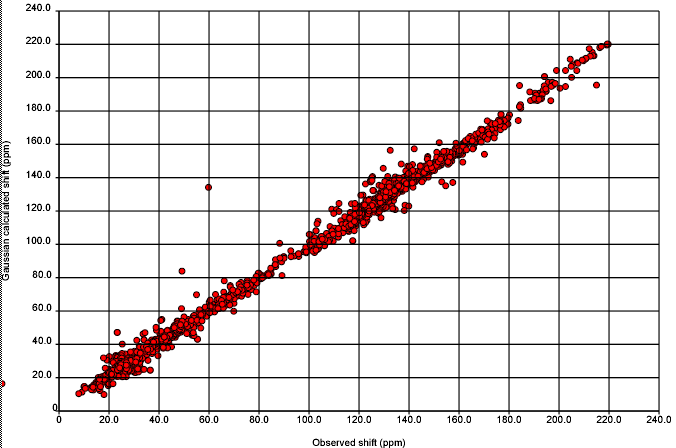
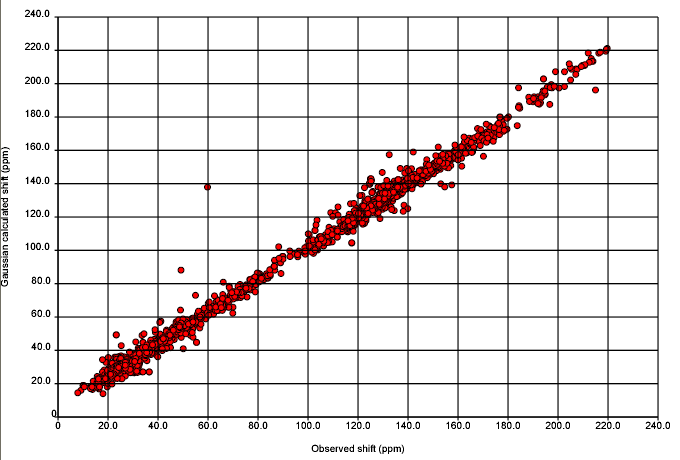
October 23rd, 2007 at 12:52 am ePeter, all that is needed to perform the calculations for comparison using the ACD/Labs NMR predictors is a download of the exact dataset Christoph provided to you (we have already had issues with comparing algorithm to algorithm but using different versions of the NMRShiftDB database…not good). Also, if Nick can send us the ID of the structure inside the NMRShiftDB this should be enough. Thanks