Prompted by an enquiry today [below] it’s a good time to update about the project. I haven’t talked to Nick today but he mailed earlier and I’ll quote from his mail, which we hope to post in full when we meet. In general from now on we’d like to make the info as public as possible.
Brent Lefebvre Says:
November 6th, 2007 at 2:49 pm e
Hi Peter,
Have you come to a conclusion? I feel that most of the responders to this blog posting have been supportive of the initiative. Please know that what Ryan Sasaki says is true; we are interested in quality science and furthering this where we can. The reason I offered to help is two-fold. We are interested in how our NMR spectrum prediction algorithms compare to the calculation methods you are proposing to use. By using this dataset as a benchmark, I think we can help this project. And if we can help this project, I hope that helps this scientific discipline, if only modestly.
Of course, our motives are not entirely altruistic. By having this benchmark test performed, we get to see how our prediction algorithms compare to the ones you are using. This can then tell us where we need to go and improve our prediction quality, which is of course, very valuable information.
I also have no problem providing you with the results of the predictions, but with the software capable of providing you with the results independently. This should assuage any fears you have of overfitting the data.
Please contact me directly if you wish to puruse this direction. We would be very happy to help.
Sincerely,
Brent Lefebvre
NMR Product Manager
Advanced Chemistry Development
PMR: We are happy to make the dataset available, but please be aware of its limitations which may disappoint you. Let me recap the design of Nick’s project which we managed to stick to:
- Could we take a heterogeneous dataset of NMR shifts and investigate whether a computational method was suitable for predicting the NMR shifts? In doing this we were aware that we would have to separate experimental variance from errors in the predictive method. When we started we had no idea of the balance between these. It could be that the experiment was very well defined and the computation poor. Or any other proportion.
- Our ultimate goal is to see whether the chemical literature as published can be analysed in this way. Could the algorithm detect errors in shifts and their assignments? If it could it would act as a robot validator for NMR during the authoring and publishing process. We have already done this for some of the data with the experimental data checker that we developed with RSC sponsorship – we wished to extend this to NMR.
- Henry Rzepa had already developed the methodology, based on work by Scott Rychnovsky and he was keen to try it out. We had already shown that the method worked for validating crystallography and it was technically straightforward to adapt it to NMR.
- We needed a set of experimental shifts with assignments. We work closely with Christoph Steinbeck AND because his database emits CMLSpect including 3D coordinates and assignments we can automatically convert the data into Gaussian input files. I repeat that I personally had no experience of the data within NMR shiftDB (and I certainly had some surprises).
Here are some excerpts from Nick’s account (I won’t post it all until I have checked that it’s OK with him). The full account has many hyperlinks and interactive displays so it’s worth waiting for:
We started off by downloading the CML files for all molecules in NMRShiftDB from here. From these we selected molecules that matched the following criteria:
- must only contain the atoms H, B, C, N, O, F, Si, P, S, Cl, Br, I
- must have MW < 300
- must not have a chain of more than two non-H atoms
- must have at least one CMLSpectrum for 13C NMR where:
- solvent is provided
- number of carbons in the spectrum must be equal to the number in the structure.
… and …
… we then attempted to find structures/assignments that were in error.
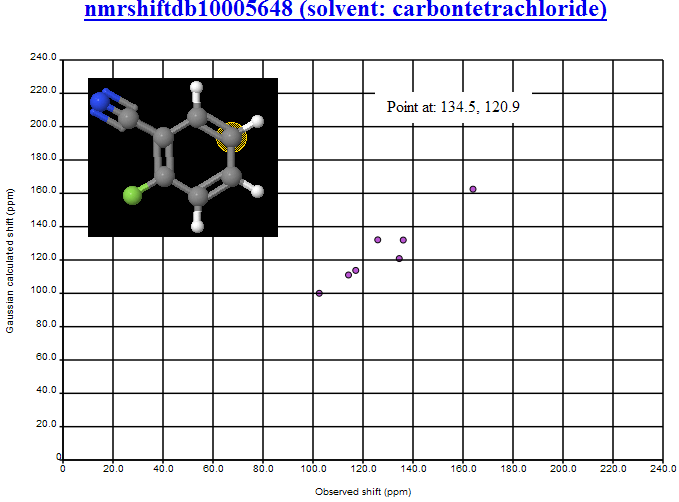
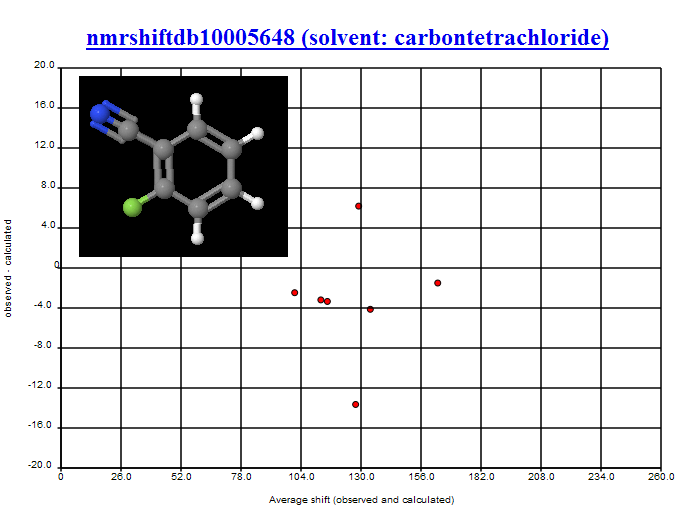
- The two structures with highest and lowest C values [PMR: offsets from TMS] are tautomers, and on investigating further with Christoph Steinbeck, we discovered that these structures had originated from the same NMR experiment, the peaks having been incorrectly manually separated before being deposited in NMRShiftDB. [PMR: This was human editing, which introduced massive errors]
- The structure with highest RMSD was highlighted on PMR’s blog, and through community discussion (notably with Wolfgang Robien and ‘hko’ (blog commenter name) it was decided that this structure was most likely incorrect.
- It was pointed out (by Wolfgang Robien) that judging from the paper for nmrshiftdb2562 it was likely that the structure given was incorrect for the spectrum.
- nmrshiftdb10008656 and nmrshiftdb10006416 we judged to have peaks that had been misassigned (these have now been corrected in NMRShiftDB).
All of the structures mentioned above were removed from further analysis.
Conformations
As only one geometry has been calculated for each structure, carbons that in reality are environmentally equivalent can be calculated to have different chemical shifts . To take this into account we used JUMBO to read in the CML files, calculate the Morgan numbers for each carbon, and average the shifts for those that were equivalent.
In many cases the problem cannot be solved this way, and some sort of conformational analysis would be needed to better predict the shifts for less rigid molecules using this method. As we are on a reasonably tight schedule, this will have to be left for another time. Thus, we removed all molecules with rings of 7 or more atoms and any molecules with likely tautomers from the dataset. This left ?? molecules to analyse.
Finding possible peak misassignments
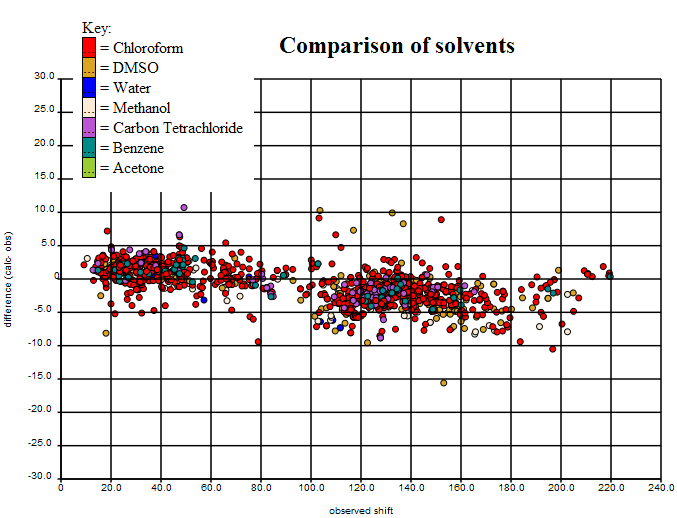
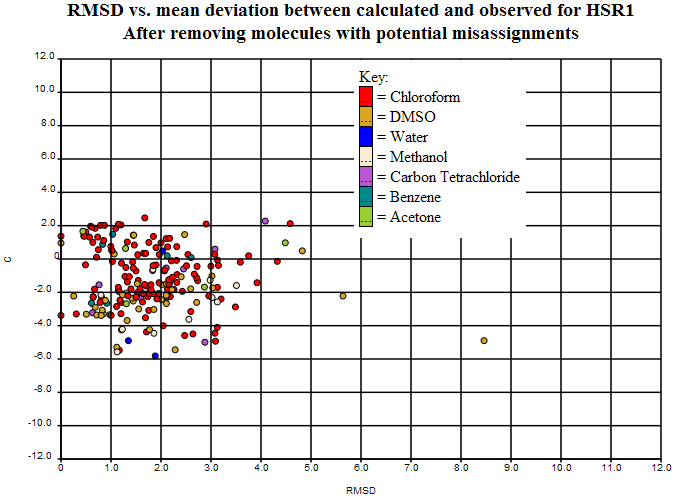
By plotting the average of observed and calculated shift against the difference between observed and calculated shift, it is possible to spot potential misassignments by looking for two points with the same x-values. In this case we’ll only mark a structure as potentially misassigned if the two peaks in question are greater than 2ppm apart. We ran a simple program over the dataset to pick out potential misassignments, and of the 249 structures left, it came up with 42 potential misassignments. The list of these (with their associated ‘misassignment’ plots) can be viewed [… tomorrow …].
Removing the potential misassignments leaves us with 207 structures,
PMR: We shall expose all data to the community. What is clear is that apart from the (expected) problems of halogens all the major variance seems to be due to experimental error. (Joe Townsend found exactly the same for crystallography – the QM methods are good at calculating geometry and good at calculating 13C).
So we are left with 207 structures that may be of good enough quality to start showing “real” variance. We know some of that is “chemical” conformations, tautomers, etc. We would expect that some is due to experimental error that is not easily identified. Some will probably be due to solvent. So only after removing these will we have a data set which is good enough to test predictive methods.
IFF, and I’d welcome the community’s comments, the data set is felt to be relatively clean then it could be a useful tool for comparing methods. But it’s relatively small and it’s probably still got errors in. If the errors are significant then it may not be a very useful test between methods.
My guess is that HOSE/NN and GIAO address problems in different ways:
- HOSE/NN will be easier to use on structures that have a number of low-energy conformers. It will be trained to give an average value whereas the GIAO method will give results for s single conformer (unless we deliberately search conformational space).
- HOSE/NN may adjust to tautomers (I don’t know) whereas if the “wrong” tautomer is chosen for GIAO it probably won’t give good agreement.
- GIAO will be better for rarely observed systems as it should be roughly independent of the atoms and their bonding.
- All methods may have virtues and defects in modelling solvation.
- GIAO is potentially capable of showing systematic errors in the data that would be absorbed by HOSE/NN methods
So when I have talked to Nick we’ll make the data and its navigation public. But the primary effact will still be asking the communitty to annotate the outliers, probably as errors. And the only allowed approaches are:
- revisit the original literature and agree it’s faulty
- write to the authors and get clarification
- create a consensus among NMR experts that a particular structure “must be wrong”.
This is a painfully slow (but necessary) way of building up a test data set. It’s what we have to do in natural language processing where each paper is hand-annotated. So if anyone has already done some of this and has a data set which is validated and Open it would be a very useful tool. The problem for GIAO is that – initially – we need fairly rigid molecules (though I think we can extend in the future).