I have been pointed to a paper in PLoSONE on Data sharing http://www.plosone.org/article/info:doi%2F10.1371%2Fjournal.pone.0021101
I haven’t read the text but I am afraid I have to comment adversely on the way the data are presented. Because it illustrates a fundamental reason why data cannot be shared. This is data from Table 6 in the paper:
This is described as “a Powerpoint-friendly Image”. It’s unreadable to a human (though if you step away it gets possible).
What does it represent? The paper describes a survey carried out by questionnaire (survey instrument) and the results are presented in 29 Tables. The entries in the tables are small amounts of text, and numbers. Here’s Table 6;
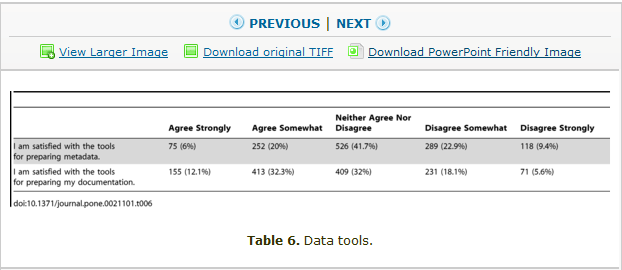
So this is a table of data. And it is transmitted as a TIFF. And if you want a “powerpoint friendly image” it appears to be a PNG.
In simple terms this completely destroys data.
Now I know some of the people involved – Carol, Cameron Neylon (who edits this, and has a journal on reproducible computing) and the folks at PLoS.
Something has gone terribly wrong here. Maybe in the authoring, maybe in the reviewing, maybe in production.
Open Access by itself solves a bit, but not enough. We also need to make our Open products BETTER than previously. Any of CSV, HTML or even XLS would be possible for the tables. Then the data could be shared.
We have to move towards fully interoperable semantic Open data.
I’m afraid the rendering of the table as a bitmapped image is an artifact of their rendering system. If you download the file as an XML, you can get the data as an HTML table (not perfect, but better than an image). Just open the XML in a text editor and search for \table 6\.
Fortunately, the authors did deposit their data in Dryad (http://dx.doi.org/10.5061/dryad.6t94p). Though, the paper was already in press, so there is no link to the Dryad data from the article.
Peter,
you can use the XML version, it has table 6 coded in XML,
machine readable, no semantics destroyed.
The XML version is next to the PDF button, however I agree
for chemistry papers there is currently no functionality at all in
PLOS publications, compared to RSC, ASC or Elsevier.
Cheers
Tobias
Thanks, Peter clearly missed that XML format. The detail in the XML seems very good to me. The PDF version also displys a good deal of metadata, but there are no links in it to downloadable XML for the tables. Maybe that is something that will be added as a result of this. I too haven’t read the paper yet, but now I have been pointed to it I surely will. Thank you.
Thanks Andy and all,
I didn’t miss the XML button, I just assumed it wasn’t worth following.
I have now looked at my own paper in PLoS and seen that they carry out the same semantic destruction.
So it’s PLoS’s fault.
This is bizarre, unnecessary and seems to require additional work – all for the purpose of making an unreadable image “Powerpoint Friendly”
I shall blog this later
Each table has a DOI, so in the PDF rendering I see doi:10.1371/journal.pone.0021101.t006 for table 6. In the html there is this link to Table 6: http://www.plosone.org/article/slideshow.action?uri=info:doi/10.1371/journal.pone.0021101&imageURI=info:doi/10.1371/journal.pone.0021101.t006
It would be good to be able to just get the table as XML or other non-image formats from that link. I suppose there could be more than one XML for the table. It’s true context is the article, so it would be good if the table XML linked back to that.
Anyway, I think PLoS One are doing a good job and driving improvments in scholarly communication. Chemistry seems to be doing better than geography in this respect, but there is a growing realisation that most of us are really doing geography of some stripe 😉
It is easy to find the XML for the table in the article XML and it has an XLink so there is perhaps really very little to find issue with about this. Perhaps the enhancement wanted is to add buttons for XML (small, medium and large) i.e. XML Table Values Only, XML Table with Metadata and context links, XML for the article. Perhaps also there could be a download package for all this as zip, tar.gz etc…
Andy,
I disagree. It may be easy for you to find the table and for me (as a practitioner of XML for 15years). But it’s completely opaque to 99% of readers.
Unless we are looking at different XLinks the one in the XML table simply links to a graphic (see its mimetype). There is no independent way of extracting the table in semantic form.
As a reasonable comparison BMC expose tables in whatever the authors sent, usually XLS.
The link you give is to a series of images, some of which (“the powerpoint friendly ones”) are unreadable as they are small PNGs. I think I am conditioned by other publishers who when they present at table provide it in more useful formats. After all it’s only an XSLT tranformation that the publisher should be able to do trivially. Whereas *we* have to work out the DTD.
Web browser rendering of the table 6 data from the XHTML 1.0 Transitional found via the following URL is good:
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3126798/table/pone-0021101-t006/
It is good because it lends itself to copying and pasting of values. The source of the file also provides the data in an XHTML 1.0 Transitional Table. However, there is no Download as XML for the table, like there is not at the other link. And for another matter XML is not the only format that others may want (like images aren’t either).
The URL for an XML file that also contains the data as values is:
http://www.plosone.org/article/fetchObjectAttachment.action?uri=info%3Adoi%2F10.1371%2Fjournal.pone.0021101&representation=XML
This is linked from the article XHTML 1.0 Transitional from PloS One as available via the following URL:
http://www.plosone.org/article/info:doi/10.1371/journal.pone.0021101
Yet, I do not see a link to an XML for the article in the similar place from PMC XHTML 1.0 Transitional available via the following URL:
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3126798/
Anyway, I think I do have an issue with pictures of a rendering of the data that do not link to the source data, but this detatchment is something that can happen. I’ve not looked at the image data and if there is any metadata that links to the data source…
Thanks very much Andy,
This confirms that if publishers care about readers it is fairly easy to make the information available in the right manner. It’s interesting how much more advanced PMC is than the journal it “abstracts”
Ok, so I was both editor and referee on this paper so to the extent that there wasn’t already a link to Dryad that I insisted on prior to publication I’ll hold up my hand as obviously not having insisted on it. I think my thought process was probably that as the data was in the paper it wasn’t necessary to push for it to be deposited elsewhere but I’d have to dig back into the editorial system to reconstruct the correspondence and that will take more time than I have today.
FWIW the review process is done entirely via PDFs so it is not straightforward to tell what the native format of any part of the paper is for the reviewer. I would agree that this is bad but its consistent with most of the journal systems I’ve worked with. Obviously this makes reviewing for data formats difficult or impossible but what’s probably worse is that it discourages you from asking the question. In some ways the BMC system is better in this respect because the files are there in front of you with big icons saying what the format is.
However I think I disagree with Peter about the destruction element here. The html version of the paper is explicitly designed in the PLoS system for human reading (admittedly by sighted people). I actually find that floating window and the ability to click through figures very useful and I’d imagine that it makes that process simple if everything, figures and tables, are the same format. Given that the tabular data is available in the XML, which is where you’d go to dig out data, I don’t think its a question of destruction but of differing priorities.
The person who wants to cut and paste the numbers from the table is going to be annoyed but the person who wants to grab the figure and drop it into a presentation is going to be happy. And I suspect the latter may be the more common re-use case. The ideal would obviously be to have both, contextually presented depending on what the user (human or machine) wants. PLoS have focussed very hard on making their html rendering attractive to human readers and have as a result pulled into a situation where html downloads are much greater than pdf downloads which I would see as a good thing. The price, with limited resources, is things like this which are suboptimal obviously.
What would a system look like that achieved all of these goals – presenting the easily cut and pasted whole for those who wanted it, plus the cut and pasted data for the humans who want that, plus the marked up data for those who want that? At least there’s a DOI for each element so a content negotiation scheme would in principle be possible. It also re-raises the question of standardising the form in which a paper points to its data on an external service such as Dryad – how should that link be made machine discoverable in a general way?
Cameron,
Thanks for the reply
>>… there wasn’t already a link to Dryad that I insisted on prior to publication I’ll hold up my hand as obviously not having insisted on it. I think my thought process was probably that as the data was in the paper it wasn’t necessary to push for it to be deposited elsewhere but I’d have to dig back into the editorial system to reconstruct the correspondence and that will take more time than I have today.
From an initial glance the Dryad deposition (which is excellent) is not the summary data reported in the paper but the raw data.
> What would a system look like that achieved all of these goals…?
It’s called PMC: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3126798/
😉
Seriously, though, tables are extremely hard to get right in all their different manifestations. I think PLoS, here, has done a great job.
This is exactly what PLoS should be doing – in fact all publishers should. So why aren’t they. Or should we get rid of publishers for dissemination (i.e. use them only for review) and use PubMed?? Makes life much easier. BTW I am on UKPMC and this gives me some ideas
Users of journals and the data they contain in articles are not just human readers that manually cut and past or download the data. There should be ways to feed and link the data that are more flexible and automatable. It is good to argue about which service does this best at and how all services can improve and standardise. It is also good that there are organisations like openresearchcomputation (http://www.openresearchcomputation.com/) which invite submission from across the boards.
Thanks also Peter for entertaining this all on your blog which is enlightening.
Maybe one day I will get around to looking at the image format data of the tables and I might be happily suprised to see the metadata I would want in them…
Thanks for your encouragement,
FWIW I am on the editorial board of (http://www.openresearchcomputation.com/)