There is some doubt about what the structure of diazonamide A is. Because there is no absolute way of assigning names to structures. We only agree what aspirin is because everyone has been assigning the same structure to it for 100+ years. Many people are careless with names and even more are careless with structure diagrams. Indeed there seems to be a minor industry in drawing some structures wrongly. A year of two back when Nick Day was pioneering the use of InChI he used “staurosporine” as an example. He found lots of structure diagrams and I think there were 19 (sic) different diagrams. Some were frankly “wrong”. Others missed out the stereochemistry, others had other problems. And some of these were from suppliers sites (i.e. “labels on bottles”).
So how can we be sure? It needs an authority – but which one? Staurosporine is a (potential?) drug, so… WHO drugs? British National Formulary? US National Pharmacopeia? Chemical Abstracts? Beilstein? All of these are pay-to-view. So I cannot look them up (remember I am at home, simulating an interested person, such as a patient). Ah! Pubchem… with 16 entries, and several variations of stereochemistry. Wikipedia has a nice picture … but this about diazonamide…
On TotSynth’s post there’s a link to the latest paper (DOI: 10.1021/ja0744448). And following this I find:
PMR: The Blue Eye is NOT part of the abstract – it shows that the Blue Obelisk Greasemonkey has found a crystalEye entry which looks like this:
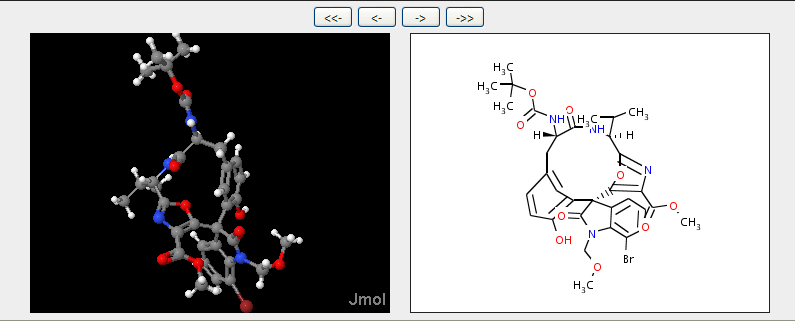
and here you can see the actual stereochemistry of the diazonamide nucleus (it’s not exactly the title compound) so there is virtually no doubt. The diagram on the right is calculated from the 3D coordinates and the layout is through CDK – note the stereo wedges and hatches.
So now I know what some of the stereo is. And because PNAS have made the text Open I can read how it relates to TS’s structure. The CDK may not be 100% beautiful, but it should be true (Cue some reader finding it’s wrong and a bug in JUMBO, but that’s what Open science is about). And you can always pay Chemical Abstracts 6.20 USD to check whether you have got it “right”.
So install the Blue Obelisk Grease Monkey (blog post) in your Firefox browser and Open your Eyes to a whole new world of truth and beauty.
-
Recent Posts
-
Recent Comments
- pm286 on ContentMine at IFLA2017: The future of Libraries and Scholarly Communications
- Hiperterminal on ContentMine at IFLA2017: The future of Libraries and Scholarly Communications
- Next steps for Text & Data Mining | Unlocking Research on Text and Data Mining: Overview
- Publishers prioritize “self-plagiarism” detection over allowing new discoveries | Alex Holcombe's blog on Text and Data Mining: Overview
- Kytriya on Let’s get rid of CC-NC and CC-ND NOW! It really matters
-
Archives
- June 2018
- April 2018
- September 2017
- August 2017
- July 2017
- November 2016
- July 2016
- May 2016
- April 2016
- December 2015
- November 2015
- September 2015
- May 2015
- April 2015
- January 2015
- December 2014
- November 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- February 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- August 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- May 2007
- April 2007
- December 2006
- November 2006
- October 2006
- September 2006
-
Categories
- "virtual communities"
- ahm2007
- berlin5
- blueobelisk
- chemistry
- crystaleye
- cyberscience
- data
- etd2007
- fun
- general
- idcc3
- jisc-theorem
- mkm2007
- nmr
- open issues
- open notebook science
- oscar
- programming for scientists
- publishing
- puzzles
- repositories
- scifoo
- semanticWeb
- theses
- Uncategorized
- www2007
- XML
- xtech2007
-
Meta
Just fyi Peter…you had commented/complained last night about the fact that we were not yet supporting 3D displays on Jmol. I quote “Now the molecules are three-dimensional but the coordinates in chemspider are those of the 2-D diagram. Personally I regard this as extremely misleading and would NEVER use Jmol for 2D diagrams, but I shan’t pursue this here.”
The molecules are NOT three-dimensional. They are 2D representations with hash and wedge bonds only. Just open the structure into any editor and you will see they are planar. The structures are not stored in the DB as 3D coordinates.
However, since this was a concern for you we’ve put smi23d under Jmol now…take a look at:
3D Optimization Now Embedded Underneath JMol on ChemSpider
http://www.chemspider.com/news/?p=79
Keep that constructive criticism coming…if you could send it directly it would make it much easier than reading all of your blog postings. You are very prolific 🙂
I’m not sure what you mean by the comment “Because there is no absolute way of assigning names to structures.” Systematic naming is exactly that….IUPAC Naming, CAS Naming. Well defined rules. Now, are they exhaustive across all forms of chemistry..surely not…inorganics, organometallics, polymers while challenging do have nomenclature standards too while some believe they don’t. Of course chemical structure classes change…there were no rules of fullerenes before they were synthesized. But, in general there IS an absolute way of assigning the names to structures. Maybe I misinterpreted your comment?
(1) … nice to get some comments
Just fyi Peter…you had commented/complained last night about the fact that we were not yet supporting 3D displays on Jmol. I quote “Now the molecules are three-dimensional but the coordinates in chemspider are those of the 2-D diagram. Personally I regard this as extremely misleading and would NEVER use Jmol for 2D diagrams, but I shan’t pursue this here.”
The molecules are NOT three-dimensional.
PMR: I know that… 🙂
They are 2D representations with hash and wedge bonds only. Just open the structure into any editor and you will see they are planar. The structures are not stored in the DB as 3D coordinates.
PMR: Agreed.
However, since this was a concern for you we’ve put smi23d under Jmol now…take a look at:
3D Optimization Now Embedded Underneath JMol on ChemSpider
http://www.chemspider.com/news/?p=79
PMR: The point is that there is an implicit assumption that if something is rendered in Jmol it is a 3D structure. It is highly likely that students and non-chemists will attempt to use such material in 3D situations. The ultimate fault is that the chemistry community do not worry about labelling a structure as 2- or 3-D. It’s often impossible to tell what a MOL file is meant to be. Of course CML overcomes all of this, but Chemspider doesn’t seem to use it.
Keep that constructive criticism coming…if you could send it directly it would make it much easier than reading all of your blog postings. You are very prolific 🙂
PMR: The postreferred to 6 different sites. I don’t have time to copy each paragraph into each of theose sites. In any case the blogosphere does that well. For example when you reference this blog we get pings/trackbacks and I read them. Works the other way as well.
I was interested in checking staurosporine on ChemSPider. Just fyi…9 hits
http://www.chemspider.com/Search.aspx?q=staurosporine
1 of these has the incorrect MF (up a OH group_ so i have curated the synonym association out.
7 have the same structure layout and 1 is different. Of the eight different structures the difference is, as usual, in the stereo layer. We are presently working to provide a visualizing solution to the connectivity diagram versus the stereo diagram(s)