I wrote a rant about PDF destroying scientific information (it keeps me sane). PDF is a hamburger. Power corrupts and Powerpoint corrupts absolutely. But they aren’t the only ways.
The third most common method of textual destruction of science is to use wordprocessors.
This is a long post. It’s longer because you HAVE to read Joel Spolsky’s post (from which I pinched the title). But you have to read it.
Word, Open Office, LaTeX. They all destroy science. Word is the worst because it’s the most commonly used and the smartest. Which means the most evil. (Yes, I’m sponsored by Microsoft to develop chemistry in Word. And those MS folks I collaborate with will understand and sympathize). Open Office will try to emulate the destruction. And LaTeX creates beautiful typeset output which also destroys science.
Your probably think I’m mad. It’s catalysed by a reply from Henry Rzepa to my post: I’m going to point to Henry’s reply rather than quote as I daren’t cut and paste. You’ll see why (and it’s not copyright this time): Here’s his comment http://wwmm.ch.cam.ac.uk/blogs/murrayrust/?p=2660&cpage=1#comment-493508. Read it carefully and tell me what you see close to the last two “ROA” strings.
I see:

If I cut and paste this I get:
the (otherwise relatively unremarkable molecule) were rather larger than normal. The relevant field is identified in the output with the string ROA– and the numerical value is identified as -9999.9. Well, for our molecule, this ended up as ROA—10000.0 (the numbers are fictitious, to illustrate the problem). You can see how one missing space totally messed.
So everything looks OK? Yes?
If, however, I use the edit function on Henry’s post I get the raw text:
intensities for the (otherwise relatively unremarkable molecule) were rather larger than normal. The relevant field is identified in the output with the string ROA– and the numerical value is identified as -9999.9. Well, for our molecule, this ended up as ROA—10000.0 (the numbers are fictitious, to illustrate the problem). You can see how one
which renders on my machine as

What has happened?
WordPress (and this would also be true of Word and Open Office) is smart. It knows that Henry “wanted” to type the em-dash character . Now it’s not on Henry’s keyboard, but WordPress (and the other corrupting processors) know what is best for Henry. It’s not what Henry actually wants, it’s what they are going to give Henry whether he likes it or not. So he types three minuses in succession ‘-‘ ‘-‘ ‘-‘
ROA—10000.0
(rendered on my machines as 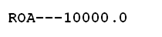 )
)
He wishes us to interpret this “R” “O” “A” “-” “-” followed by “minus ten thousand”.
[Note incidentally that as I type this, Word has CHANGED, yes CHANGED, the quote character (ASCII 34) in my text to the evil smart quotes – the sloping things. Word knows what’s best for me as well as for Henry.]
So whereas Henry wrote something perfectly sensible and important, the tools have changed his text to gibberish.
THE ONLY FORMAT THAT SHOULD BE USED FOR TRANSMITTING SCIENCE IS ASCII. Code points 32-127.
Help! What’s a code point?
A code point is the platonic identification of the character. Not what it looks like (its glyph). Not what it is transmitted as (its encoding). The character itself.
So If we can only use code points 32-127 how do we do important things like the degrees sign? And the world-conquering copyright symbol? They aren’t in ASCII.
YOU MUST NOW READ JOEL SPOLSKY. The Absolute Minimum Every Software Developer Must Know About Unicode and Character Sets
And Wikipedia: http://en.wikipedia.org/wiki/Unicode .
If you do not understand the difference between code points, encodings and glyphs, and if you are creating any form of data-rich scientific document then you are probably committing a crime against science.
You are randomly multiplying your numbers by -1.
You are randomly destroying symbols denoting units
You are randomly losing digits.
You are transforming “mu” to “m”.
I’m sorry, but there is no escape. You HAVE to understand lab safety, right? It’s part of science.
So is character encoding.
So here is the good news:
When every scientist and scientific publisher uses Unicode, and when all the toolsets support Unicode and when documents are transmitted with a clear encoding (such as UTF-8), then the problem goes away.
And the bad news is that we are not there yet:
Publishers produce gibberish on their web pages. (Not all pages and not all publishers, but there is still gibberish). Here’s one of the good (the Royal Society of Chemistry):
<!DOCTYPE html PUBLIC “-//W3C//DTD XHTML 1.1//EN” “http://www.w3.org/TR/xhtml11/DTD/xhtml11.dtd”>
<?xml version=”1.0″ encoding=”utf-8″?>
<html xmlns=”http://www.w3.org/1999/xhtml” xml:lang=”en”>
It specifies the XHTML namespace, the human language (EN) the DTD (for validation) and the ENCODING (UTF-8). Everything that should be there.
And here is the ACS:
<!DOCTYPE html PUBLIC "-//W3C//DTD XHTML 1.0 Transitional//EN"
"http://www.w3.org/TR/xhtml1/DTD/xhtml1-transitional.dtd">
<html xmlns="http://www.w3.org/1999/xhtml">
Missing the language and the encoding. But at least they got the COPYRIGHT properly encoded. It wouldn’t do to fail to display that:
<div><a href="/action/clickThrough?id=317672&url=http%3A%2F%2Fportal.acs.org%2Fportal%2FPublicWebSite%2Fcopyright%2Findex.htm&loc=%2Fjournal%2Fjoceah&pubId=40026046" title="ACS Copyright Information" class="underline">Copyright © 2010 American Chemical Society</a></div>
Where's the copyright symbol? It's the "©" See Wikipedia: http://en.wikipedia.org/wiki/Copyright_symbol which says:
The character is mapped in Unicode as U+00A9 © copyright sign (HTML: © ©).[4] On Windows systems, it may be entered by means of Alt codes, by holding the Alt key while typing the numbers 0169 on the numeric keypad. On Macintosh systems, it may be entered with ⌥G. The HTML entity is ©, and it can also be referenced as © or ©.
Unicode has also mapped U+24B8 Ⓒ circled latin capital letter c and U+24D2 ⓒ circled latin small letter c.[5] They are sometimes used as a substitute copyright symbol where the actual copyright symbol is not available in the font or in the character set, for example, in some Korean code pages.
But hang on! I've just copied those paragraphs into TextPad and what I see is:
The character is mapped in Unicode as U+00A9 ©? copyright sign (HTML: © ©).[4] On Windows systems, it may be entered by means of Alt codes, by holding the Alt key while typing the numbers 0169 on the numeric keypad. On Macintosh systems, it may be entered with ?G. The HTML entity is ©, and it can also be referenced as © or ©.
Unicode has also mapped U+24B8 ?? circled latin capital letter c and U+24D2 ?? circled latin small letter c.[5] They are sometimes used as a substitute copyright symbol where the actual copyright symbol is not available in the font or in the character set, for example, in some Korean code pages.
It’s completely trashed. AND THIS IS WHAT HAPPENS WHENEVER YOU CUT AND PASTE INFORMATION WITH TOOLS THAT ARE NOT UNICODE COMPLIANT AND WHERE THE ENCODING IS NOT SET. Notice that many of the symbols are replaced by question marks (?). And the small capitals have been turned into lower case. And goodness knows what else.
Is it Wikipedia’s fault? Possibly. Their page shows:
<!DOCTYPE html PUBLIC "-//W3C//DTD XHTML 1.0 Transitional//EN" "http://www.w3.org/TR/xhtml1/DTD/xhtml1-transitional.dtd">
<html xmlns="http://www.w3.org/1999/xhtml" lang="en" dir="ltr">
<head>
<title>Copyright symbol - Wikipedia, the free encyclopedia</title>
<meta http-equiv="Content-Type" content="text/html; charset=UTF-8" />
<meta http-equiv="Content-Style-Type" content="text/css" />
They have identified the charset to the HTML agent (browser) but this isn’t the same as actually adding the encoding. Let’s edit the Wikipedia page in TextPad. And we get a message:
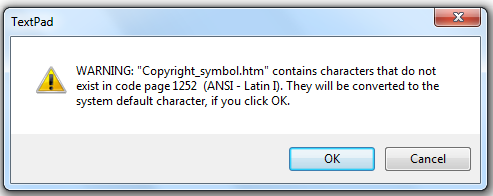
As soon as you see this you can be CERTAIN THAT YOUR INFORMATION IS BEING CORRUPTED. What it means is “You haven’t given TextPad the encoding so TextPad (which is a good program) has to guess what the characters are”.
What’s 1252? see http://en.wikipedia.org/wiki/Windows-1252 which explains:
Windows-1252 or CP-1252 is a character encoding of the Latin alphabet, used by default in the legacy components of Microsoft Windows in English and some other Western languages. It is one version within the group of Windows code pages. In LaTeX packages, it is referred to as ansinew. The encoding is a superset of ISO 8859-1, but differs from the IANA’s ISO-8859-1 by using displayable characters rather than control characters in the 0x80 to 0x9F range. It is known to Windows by the code page number 1252, and by the IANA-approved name “windows-1252”. This code page also contains all the printable characters that are in ISO 8859-15 (though some are mapped to different code points).
It is very common to mislabel Windows-1252 text data with the charset label ISO-8859-1. Many web browsers and e-mail clients treat the MIME charset ISO-8859-1 as Windows-1252 characters in order to accommodate such mislabeling but it is not standard behaviour and care should be taken to avoid generating these characters in ISO-8859-1 labeled content. However, the draft HTML 5 specification requires that documents advertised as ISO-8859-1 actually be parsed with the Windows-1252 encoding.[1]
If you don’t understand this, it means “Windows does it differently from everyone else”. But, to be fair, everyone else (nearly) is also making a pig’s ear of it.
So the simple rule is:
DON’T CUT AND PASTE NON-ASCII CHARACTERS WITHOUT AN EXPLICIT ENCODING.
Which simplifies to
DON’T CUT AND PASTE UNLESS YOU KNOW WHAT YOU ARE DOING AND YOUR KNOW YOUR TOOLS ARE COMPLIANT
Which normally simplifies to
DON’T CUT AND PASTE
So this looks pretty bleak. Many tools are non-compliant. Almost all documents are non-compliant. Almost no scientist understands the problem. People are used to seeing gibberish in their browser. In Word. In LaTeX.
But it doesn’t apply to science, does it?
Here’s a simple true sentence:
1 μg = 0.001 mg
And cut and paste reduces it to
1 mg = 0.001 mg
Well if your experiments don’t worry about a factor of 1000, then don’t worry. This is happening every day – micrograms are transformed to milligrams. A clever tool somewhere “guesses”. And of course cutting and pasting from Powerpoint almost certainly guarantees this
Yes, if you think LaTeX is never evil, try the Hufflepuff test.
How many F’s are there in this sentence? Put your cursor here and search for “f”.
Hufflepuff, my Hufflepuff!
Well it depends on what you mean by “f”. A human may see 8. A machine sees only 4. Because the code points in the first word represent ligatures. Let’s look in a preformatted (monospace) font which shows the two words are different.
Hufflepuff, my Hufflepuff!
There are 7 code points in the first, 10 in the second. The first will NOT be found by natural Language processors (unless they expand out ligatures). This is what I created in HTML:
<html>
Hufflepuff, my Hufflepuff!
<pre>
Hufflepuff, my Hufflepuff!
</pre>
</html>
(Yes, I omitted the encoding). The first word defines two Unicode code points which represent the ligatures. I doubt there is any chemical text processing in the world which treats ligatures properly. We put a lot of effort into trying to recover from em-dash instead of minus. All because the tool sets and the publishers create underspecified information.
So what happens when we paste our sentences into TextPad? You should be able to guess:
Hu?epu?, my Hufflepuff!
Hu?epu?, my Hufflepuff!
The ligatures are replaced by “?”.
So this is a long post. There is no absolute solution as the toolset and the publication process is almost universally broken. Science progresses one funeral at a time, so it will take some years. But here are some rules:
- USE XHTML with UTF-8. It costs nothing. It’s free. You don’t have to pay for it. You won’t break copyright.
- USE A UTF-8 COMPLIANT TOOL. Most good Open tools will do this.
- TURN OFF ALL SMART TYPESETTING IN YOUR TOOLS. QUOTES, MINUSES, LIGATURES.
- THINK ABOUT THE PROBLEM.
- USE MONOSPACE (COURIER) FOR IMPORTANT SCIENTIFIC INFORMATION, DATA, CODE. ARGUE WITH ANY PUBLISHER WHO TRIES TO RESET MONOSPACE INTO GARBAGE.
- WRITE TO ANY PUBLISHER WHOSE PAGES FAIL TO DISPLAY FUNDAMENTAL SCIENTIFIC INFORMATION CORRECTLY in all browsers.











 quixote-qcdb@googlegroups.com ). You don’t have to be a chemist. You just have to enjoy seeing Open scientific data.
quixote-qcdb@googlegroups.com ). You don’t have to be a chemist. You just have to enjoy seeing Open scientific data.



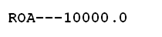 )
)