#oss2011
Jenny Molloy and I have been representing the Open Knowledge Foundation at the Open Science Summit and we presented the Open Research Reports (ORR) project. The slides we used are at http://dl.dropbox.com/u/6280676/orr.pptx. I expect that at some stage we’ll be on the video record (last year’s was very useful and also there was a transcript!). Because what we say affects the understanding of the slides.
The slides came from several sources:
- The presentation by David Shotton and Tanya Gray at Science Online this September in London. ORR arose from ideas from David and others of us who met at “Beyond the PDF” where the idea of ORR emerged (idea don’t belong to people, they choose people). The SoLo presentation gave lots of detail on the Semantics, which wouldn’t have fitted into a 13-minute slot, but the WHY slides and some of the WHAT and HOW were included.
- Jenny’s overview of the London discussion and our further groundwork with JISC, OKF and SWAT4LS. See http://www.ukoln.ac.uk/events/devcsi/life-sciences-hackdays/index.html
- My thoughts on the WHY of Open Research Reports (many expressed in animalophoto-comics)
- Jenny’s overview of what we’re going to do in ORR and particularly the Hackathon in December
- Results of the Open Bibliography/Citations projects
The slides themselves tell only part of the story – what follows is my thoughts alone and (probably) what I said. I was somewhat provocative and any flak should be directed at me, not Jenny, David or Tanya.
Summary:
Open Knowledge saves lives
ORR is A community project to make disease data Open
We started with the (obvious) truth that information is a key component of health-care. That it’s critical for the poorest countries in the world. So isn’t it already catered for by the HINARI program http://en.wikipedia.org/wiki/HINARI which “was set up by the World Health Organization and major publishers to enable developing countries to access collections of biomedical and health literature.”
So the publishers make their electronic material freely available (presumably gratis not libre) …
The country lists are based on Gross National Income (GNI) per capita (World Bank figures). Institutions in countries with GNI per capita below $1600 are eligible for free access. Institutions in countries with GNI per capita between $1601-$4700 pay a fee of $1000 per year/institution
So isn’t this very commendable of the publishers to give their material freely to those most deserving? And when the countries become rich , they can pay. Well, this year Bangladesh became a richer country and the HINARI journals were cut off. There was outrage, reported by the Lancet (itself an Elsevier journal and closed access so presumably the Bangladeshis couldn’t even read the outrage). Read http://download.thelancet.com/flatcontentassets/pdfs/S0140673611600664.pdf – which appears to be gratis. And the LANCET argued that the HINARI should be re-extended to Bangladesh.
But I think that’s completely wrong. The HINARI program only exists because the publications are CLOSED. It costs nothing to make the journals available. It costs more technically to prevent people reading the literature than to make it available. Libre material gets copied at zero cost. HINARI is nothing more than the crumbs of charity that the kinds used to give out. HINARI perpetuates a morally unacceptable system. The publishers aren’t giving their content free, they are giving OUR content free (or rather restricting access to our content).
Simply, closed access publishers make money by restricting access to information.
That’s been a consistent theme through the discussion
Now we all agree, I think, that more and better information leads to better medicine, better health-care, better environment.
And
The worse the medicine and healthcare, etc. the more people die.
Nothing controversial so far? But these are the premises of a syllogism, and when followed through you end up with the conclusion:
Closed access means people die
I don’t think anyone can deny the truth of that conclusion. If a doctor, a patient, a planner, an engineer, cannot read the appropriate literature then they make suboptimal decisions. And that means people die.
So the balance is:
If we want a closed access publishing system then we have to accept that the price is people’s lives.
Well, isn’t that how the world just is? Engineering has fatalities, Transport has fatalities, leisure sports have fatalities, so why not scholarly publishing?
Because it’s completely avoidable. The more I write about Openness the more angry I get about the immorality of closed access and walled gardens. And even more angry about the lobbying, the politics that tries to close down open efforts. We heard today (not from me) about how the American Chemical Society had spent money and lobbied to have Pubchem (the repository of Open chemical structure information) shut down. So my language is now less nuanced
Closed access means people die
And that’s not just me. In ORR we are having major contributions from Graham Steel and Gilles Frydman – patient champions for CJD and Cancer. Gilles told me of hundreds of people who die if their physicians don’t know about the latest literature. Remember these physicians cannot read the literature (there’s a blithe and stupid assumption that because they are professionals they don’t have to pay for the medical literature – the papers WE scientists give the publishers in return for our h-indexes). So misdiagnosis is common and avoidable by access to the literature. (And don’t dare try to tell Gilles he and his 65,000 community are not qualified to make this assessment).
So then we moved to WHAT can we do and how can we do it. The basic idea is to take the Open material – primarily libre material in (UK)Pubmedcentral – and collate it into annotated quality reports, one per disease. Collect the Open papers, and rank them by citation (yes I know that’s imperfect, but we aren’t trying to advance someone’s career, we are trying to save lives). Then we get the community to annotate them.
What? Get unqualified people to extract information from the papers? That’s junk.
No. here’s the sort of material that David has listed for an infective disease paper:
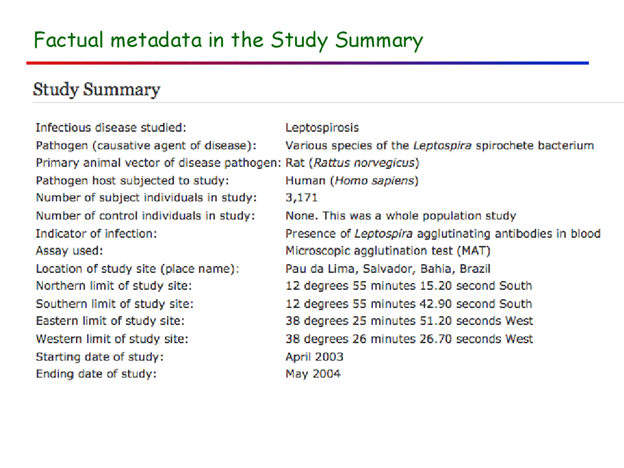
I am sure that no one reading this would be unable to extract SOME of this information reliably. You don’t have to be a medic to understand lat/long or dates or species. I personally would be able to extract all the drugs (and even our software can). So each person does what they can do well.
And as a result the community edits the report.
And we have a high-quality tool.
Hang about! You’ve omitted all the closed access stuff. That’s 90% of the literature.
So? For many purposes 10% is completely sufficient. For introductory material, for teaching, for a text-mining corpus, for diagrams, the list is endless.
And the quality of the annotation and extraction can be used for data-mining, mashups, all sorts of semantic stuff. Making it much more useful than the same amount of stuff in the closed literature.
And mightn’t this just jerk the consciences of some people? And continue to tip the balance to Open.
So join us for the Hackathon in London, UK on 2011-12-06/07. Because it’s a hackathon we don’t know in detail what we’ll do. But we’ll make a major start to establishing Open Research Reports.
And of course YOU can take part. The literature must be all Open, so everyone can read it. All tools will be Open. It’ll be great fun. Closed access publishers especially welcome as it will help them to adjust to the inevitable change taking place.

















 x
x