My unit tests have taken over my life this weekend. This post is a brief respite…
Unit tests are one of the great successes of modern approaches to programming. If written briefly about them before and Egon has an introduction of how they are used in Eclipse. Unit tests are often seen as part of “eXtreme Programming” though it’s possible to use them more or less independently. Anyway, I was hoping to add some molecule building extensions to JUMBO, and started on the CMLMolecule.class. Even if you have never used Unit tests the mental attitudes may be useful.
I then discovered that only half the unit tests for this class were written. Purists will already have judged me on this sentence – the philosophy is to write the tests before the code. For the CMLMolecule class this makes excellent sense. Suppose you need a routine:
Point3 molecule.getCentroid3()
then you first write a test. You might create a test molecule (formally a “fixture”), something like:
CMLMolecule testMolecule = makeTestMolecule();
Point3 centroid = testMolecule.getCentroid3();
Point3Test.assertEquals("centroid",
new Point3(0.1, 0.2, 0.3), centroid, EPSILON);
I haven’t written the actual body of getCentroid3(). I now run the test and it will fail the assertion (because it hasn’t done anything). The point is I have a known molecule, and a known answer. (I’ve also had to create a set of tests for Point3 so I can compare the results).
You won’t believe it till you start doing it but it saves a lot of time and raises the quality. No question. It’s simple, relatively fun to do, and you know when you have finished. It’s tempting to “leave the test writing till later” – but you mustn’t.
Now why has it taken over my weekend? Because I’m still playing catch-up. At the start of the year JUMBO was ca 100,000 lines of code and there were no tests. I have had to retro-fit them. It hasn’t been fun, but there is a certain satisfaction. Some of it is mindless, but that has the advantage that you can at least watch the football (== soccer) or cricket. (Cricket is particularly good because the cycle between action (ca 1 minute) often coincides with the natural cycle of edit/compile/test).
It’s easy to generate tests – Eclipse does it automatically and makes 2135 at present. If I don’t add the test code I will get 2135 failures. Now Junit has a green bar (“keep the bar green to keep the code clean”) which tells you when the tests all work. Even 1 failure gives a brown bar. The green bar is very compelling. It gives great positive feedback. It’s probably the same limbic pathways as Sudoku. But I can’t fix 2135 failures at one sitting. So I can bypass them with an @Ignore. This also keeps the bar green, but it’s a fudge. I know those tests will have to be done some day.
So 2 days ago JUMBO had about 240 @Ignores. Unfortunately many were in the CMLMolecule and MoleculeTool class. And the more tests I created, the more I unearthed other @Ignores elsewhere.
So I’m down to less than 200 @Ignores now. I’ve found some nasty bugs. A typical one is writing something like:
if (a == null)
when I mean
if (a != null)
This is exactly the sort of thing that Unit tests are very good at catching.
So I here’s my latest test
oh dear! it’s brown. I’ll just @Ignore that one test and…
and it’s GREEN!
(But I haven’t fooled anyone. The @Ignore is just lying in wait to bite me in the future…)


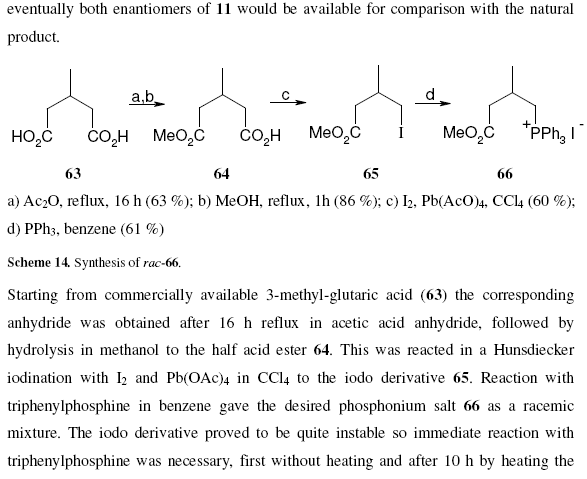
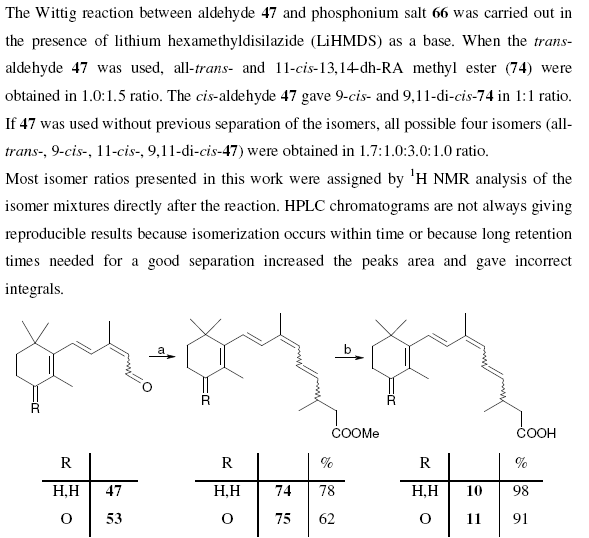
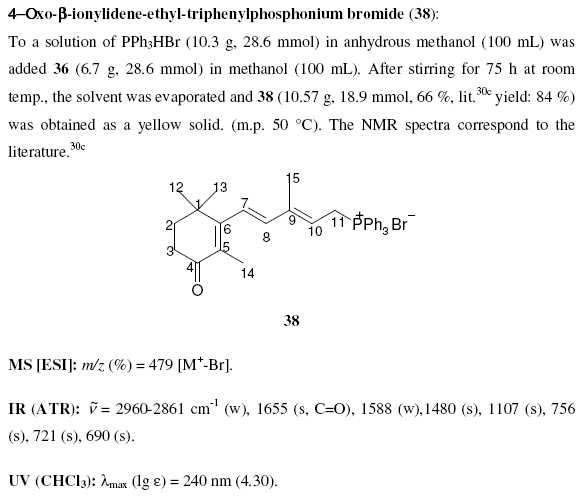
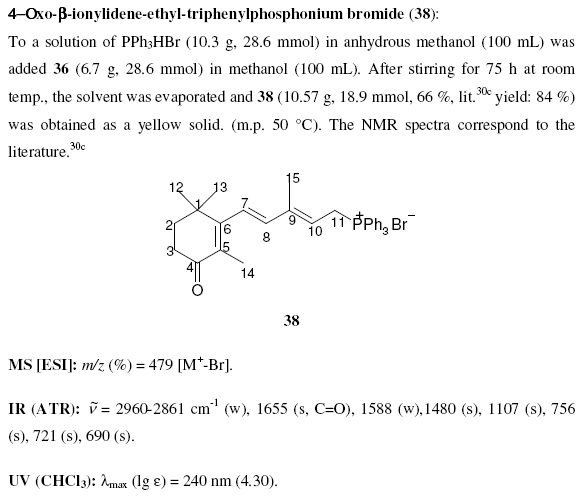


October 18th, 2006 at 10:18 am eOK – having carefully and rather too obviously written in InChI and SMILES strings in a story about ozone at nexus.webelements.info, and being an inorganic chemist who might want to write about a few inorganic species, I wondered how to write strings for, say, metal coordination complexes like the salt [Cr(OH2)6]Cl3. This compound is listed at PubChem athttp://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=104957
but shows a nonsense structure, and not being a fluent InChI reader I therefore distrusted the InChI string on that page. I looked at the above mentioned carcinogenic potency database and found
http://potency.berkeley.edu/chempages/COBALT%20SULFATE%20HEPTAHYDRATE.html
where again the chemical structure drawn is nonsense and so again I have little confidence in the InChI string on that page.
So how does one proceed for such species?