So far I have had replies from Antony Williams of Chemspider and Steve Bachrach. These are thoughtfull and I’ll comment here:
PMR: I’ll reply to the main body later, but the argument that “MANY organizations do something, so it’s appropriate” is invalid. Many organizations take my Open Access article, and resell it, some with their own copyright. This is immoral and illegal and increased number makes it worse, not better.
Much of what Antony writes deserves comment:
Posted by: Antony Williams in UncategorizedCopyright©2007 Antony Williams
Peter Murray-Rust has initiated a very LIVELY conversation about Wiley’s Chemgate rollout in collaboration with eMolecules. …
In that blog Peter posed an interesting question – “Now, Wiley publish a lot of chemistry. And they accompany this with supplemental info – data which they copyright. Do you think if I ask them nicely they will let me aggregate this non-copyrightable data in the same way as we have done for CrystalEye? Please, Wiley, let me know. ”
This is a very interesting opening for a discussion that has concerned me for a while. Specifically, many companies are aggregators of published data which is then commercialized. Most groups who build databases extract from the literature – whether these are databases of PhysChem properties, assigned NMR shifts, toxicology data etc these are commonly extracted from the literature. This has been going on for decades. if the data are copyrighted within the article then is this permissible? The data on NMRShiftDB may exist within articles and be online…permissible? The data on QSAR World are from the literature..permissible? If there are copyright issues with extracting single measured data points (e.g solubility, pKa, logP values) from the literature or even NMR assignments then the number of potential lawsuits is enormous. I don’t believe it’s possible.
PMR: This is a fair summary of the C20 situation. Many organizations extract and aggregate data. Some simply list it, some comment on it, some select or create the “best” (critical) values. All this is highly worthy. And it’s also reasonable that they should be rewarded for their effort.
But they shouldn’t use this to monopolize the use of the data – later.
What would happen now if someone chose to post a structure and a “value” extracted from a paper to the submission system on ChemSpider? Illegal?
Peter then posted a little more asking a direct question about copyrighting. He commented:
“This is an offer to Wiley (or eMolecules) to explain why they feel they are legally and morally allowed to copyright data and resell it. This blog is developing a tradition of offering publishers a chance to put their view in a highly public forum, so I would be grateful of a reply. ”
also “I realise that not all entries in ChemGate come from Wiley journals – some are private contributions, and presumably some are abstracted from competitor journals. But I would be amazed if there were not entries corresponding to Wiley journals.”
I am not aware of the details regarding the spectral collections on ChemGate in terms of copyright so I did a search online and found that the original ChemGate was probably served up by Specinfo technologies and listed the databases on there under their own branding. I believe this is a revenue sharing model with the generators of the data – there is lots of detail online regarding the providers of the data so I won’t bore you with it – however, I don’t believe any of it is extracted from Wiley publications which is what I believe Peter is pointing to. Is there any evidence that any of the data are extracted from Wiley publications? MORE comments below after the list..
[potential suppliers of info snipped…]
I believe that Wolfgang Robien’s spectral collection has likely been added to the list above (but it might actually already be a subset of the Wiley collection and licensed from Robien directly. Peter has already commented recently about NMR data and predictions directly in relation to NMRShiftDB discussions. This discussion is different since I believe this is about a spectral curve collection OR spectra reconstructed from assigned data.
My best judgment is that the data on ChemGate are likely all appropriately copyrighted. Why do I say this? If you visit ChemGate online at http://chemgate.emolecules.com/ you will see the list of NMR nuclei is : 1H, 13C, 11B, 15N, 17O, 19F , 29Si, 31P. if you visit the Modgraph site then you will see the following statement:
PMR: I have no problem about the database creator and owner owning the collection. I do have problems about copyrighting each individual spectrum since I believe that spectra are data – and therefore not copyrightable. IOW if I use a spectrum from one of these collections the copyright issue is not whether I have the right to reproduce (and potentially resell) the spectrum, but whether I am allowed to extract a certain percentage from the collection.
“The main NMRPredict program is supplied with 131,569 C13 records abstracted from the literature by Professor Robien and co-workers at the University of Vienna over the past 25 years. Three optional additional database available are:
[numbers deleted]
If you compare the nuclei in the list above, and the list on ChemGate, as well as look at the bolded statements, then I judge that ChemGate is serving up the Wiley copyrighted collections (licensed or purchased from their collaborators) rather than serving up any copyrighted data from their articles. So, I think Peter can relax about that.
PMR: It may well be that this is currently true – though I’d like to have definitive statements from the parties involved. But there is no guarantee that this won’t happen in the future and every likelihood that it will. The journals – which now increasingly require authors to publish spectra (rightly) should not immediately feed them back into the publishers databases where they are resold to the authors.
However, based on the comments made on ModGraph’s site and bolded above there may be an issue about copyrights. “In the next few years a dramatic expansion of the databases behind NMRPredict can be expected. The journals selected will cover mainly heterocyclic and medicial chemistry in order to give reliable predictions for candidates for drug discovery (Lipinski’s “Rule of Five”). ” It is unlikely that assigned spectra will be extracted only from Wiley publications despite their special relationship  .
.
I DO NOT believe that there is an issue with extracting assigned spectral data and associated structures from any publications. If there is an issue with this I am very interested in having a publisher declaring that here since I know there are parties reading this blog that do exactly that for their business!
PMR: There are grey areas here. If I sit down (or contract out to wage slaves) to transcribe data manually (with quill pen and parchment) and then type it up again then I think no publisher has an issue. If I read the spectrum electronically and put it in a database then I am liable to be pursued by the publisher for breach of rights. So we have the absurb situation that the only way we can get data is to transcribe it by hand. And each user has to do this. What a laughing stock chemistry is to the IT community.
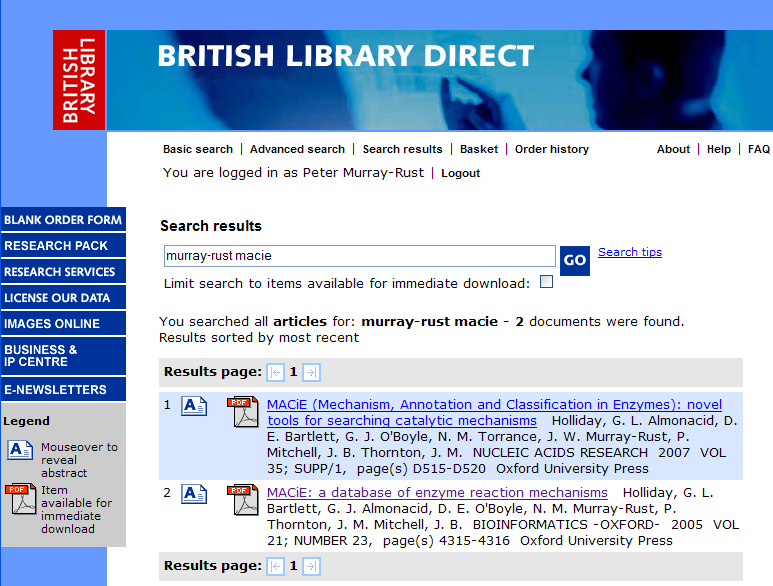
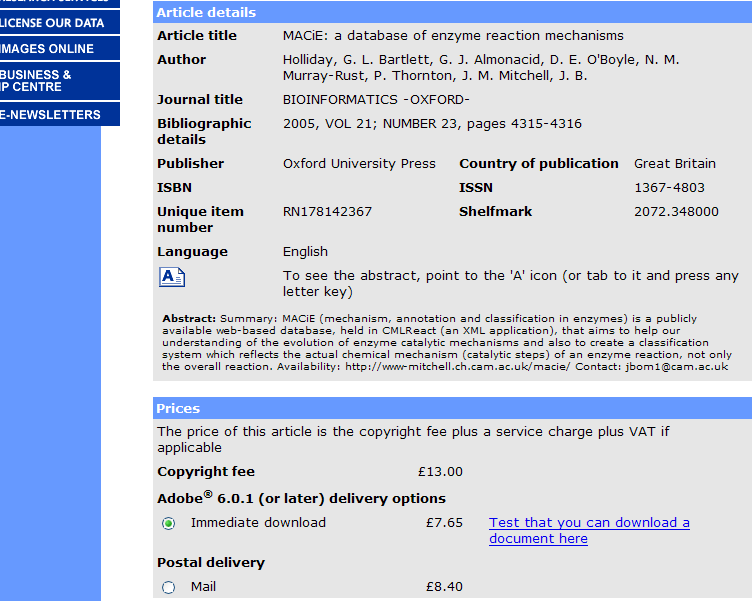
I will be contacting Wiley this week with the request to index the structures and links back to ChemGate here on ChemSpider. Having been involved with creating spectral databases over the years I believe that the pricing for access to over 700,000 spectra is actually very good. Academic prices are likely lower than those listed online.
PMR: 35,000 USD per year is prohibitive. Even if we could afford it we wouldn’t be able to do anything useful other than look at the spectra because the rights would forbid us to redictribute enhanced data.
Question for all… we are presently accepting both spectra and, shortly, structures and associated data onto ChemSpider. Question for the readers – what is the preferred Creative Commons license you would like to see attributed to the user uploaded data?
PMR: The only one that makes sense is CC-BY. Attribute the author and protect their rights. Allow anyone to re-use the data for whatever purpose including commercial. Unfortunately this doesn’t prevent the aggregation and claims to ownership. So, for example, if I publish 100,000 data and an organization integrates it into its own collection it can claim ownership to the collection. That may make it very difficult for the original author to make sure their rights are fully protected.
I think you for this genuine offer to explore this. I would suggest you contact John Willbanks at Science Commons to see if they have suggestions
Peter commented on his post “I had offered eMolecules 250 000 MOPAC calculations as Open Data.” Peter, we’d welcome the opportunity to host your data for everyone.
September 10th, 2007 at 3:50 am eI have commented in a recent blog post about your comments regarding copyrighted data extracted from Wiley articles and resold as a spectral collection. http://www.chemspider.com/blog/?p=126Bottom line…I don’t think there is an issue…all data are probably licensed/purchased from appropriate sources. Now, the issue is whether it is appropriate for an organization/individual to extract and compile literature data and commercialize. MANY organizations do this so I think it is appropriate.