A correspondent asked my opinion about Freebase:
This blog entry may be of interest, about Freebase a collaborative database project which may or may not be open. Are you familiar with it?I must admit to being rather suspicious about anything “Inc” being behind a project like this, but it certainly seems to be something to watch.
PMR: I’m familiar with some of the Freebase folks – having had drinks at WWW2007 – but I have not used it. However I believe in the concept and in a future post will detail how we are (thinking of) making crystalEye available under Freebase and other such offerings. Here, however, I catch up on a lovely phrase in the first URL…
Freebase, Wikipedia and the right to fork
Two nights ago I went to the first Freebase user meeting outside the US. (You can tell I’m setting myself up for a, “I was there when…”)
[image snipped]
It was organised by Kirrily Robert, who’s taken enough with her “new crack habit” to set up a specialised blog just for it.
So, what is Freebase? It claims to be a “database of everything”. There are several points of comparison with Wikipedia. Where Wikipedia is an “encyclopedia”, Freebase wants to be “everything”. It is far more structured than Wikipedia (which anyone who’s ever wrangled with an esoteric template might appreciate). Like Wikipedia, it’s a free content project: data derived from Wikipedia is GFDL (natch) and everything else is CC-BY. They have a very excellent and well-documented API — they’re not afraid to share. Bring on the mash-ups!
There are several more differences worth discussing. Currently, Freebase is alpha and invitation-only for write permission (ie an account). No worries, give it time.
[… details of Freebase snipped …]
So, interesting to see what will happen there. It’s Wiki[p|m]edia that convinced me (and taught me) about the absolutely vital right to fork. That is an incredible freedom which is vastly underappreciated by the journalists who are generally impressed with Wikipedia’s “freeness” (meaning no ads, or free access). And as a project leader, any kind of project, that is what keeps you on your toes. Maybe it is a good benchmark for deciding if you want to be a contributor to a particular project. If management gets too heavy, you can keep them in line by threatening to exercise your right to fork. Yeah!
Back to Freebase… another related, interesting aspect will be watching the development of their community and how it will be managed. Where Wikipedia was pretty grass-roots, it seems like Freebase is top-heavy, for the moment at least. Letting go, giving up control and trusting the unwashed masses is a very difficult psychological moment for anyone (who’s not a Wikimedian). Trying to get those same unwashed masses to behave themselves is a whole other kettle of fish. When I first contemplated this for Freebase two night s ago I was filled with cynicism, until I remembered… The thing about Wikipedia is that it only works in practice. In theory, it can never work.
I should make that my mantra. Every time I get cynical about something, think about that idea again. It only works in practice.
PMR: This is a beautiful description of the spirit (and it is spirit) of “Web 2.0”. (I am not religious about this phrase but it is a useful placeholder). I think the word mantra is apt. Only by completely releasing everything do you gain the world. If you put you and yourself at the centre you are not Web 2.0 and you will not prosper.
I learnt this about 5 years ago with JUMBO – my CML browser – and Jmol. JUMBO was Open Source, but I wanted it to do everything – 2D editing, 3D building, 3D display, 2D display, file handling, spectra, compchem, etc. And it DID all of these. It had the functionality of Bioclipse 1.0. But it used Swing (sick) and it sucked. Really badly.
So I realised that my giving up “my” possession of the 3D graphics space to Jmol, I would benefit far more than if I tried to keep everything to myself. I threw out the 3D display and bolted in Jmol – at that stage I think the Dr. Who of Jmol was Egon – it was pre-Miguel.
And so, when we Open Source chemistry hackers met in San Diego 2? 3? years ago it was natural to form the Blue Obelisk. The name chose itself – there were 2, and Geoff Hutchison spent a long time waiting at the Lesser Blue Obelisk. It’s in the spirit of TimBL’s mantra at WWW2007 – “Just Do It”. Since WP reminds us this is Nike’s slogan, perhaps JFDI is better.
So the skill – which is easy to state, but doesn’t gurarantee any outcome – includes: Do the bits you are best at. Do not care about ownership. Free them completely. You must care about freedom. Find other who add the complementary bits. Form a community.
So, for example we are exploring Freebase for chemistry and starting with crystalEye. We have 130,000 molecules with superb data and metadata. Freebase, when we learn how to use it, allows us to query on any of the metadata. Indeed it should be possible to mash this with huge numbers of other resources…
Will the chemists be interested? A few enlightened ones. But that is all it takes to start viral forks
P.
The only non-freedom is the requirement to protect freedom.
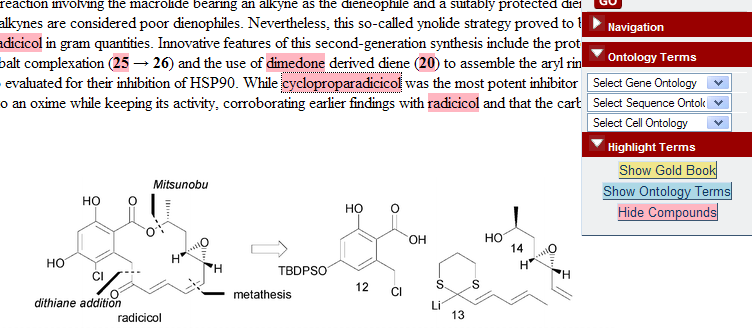
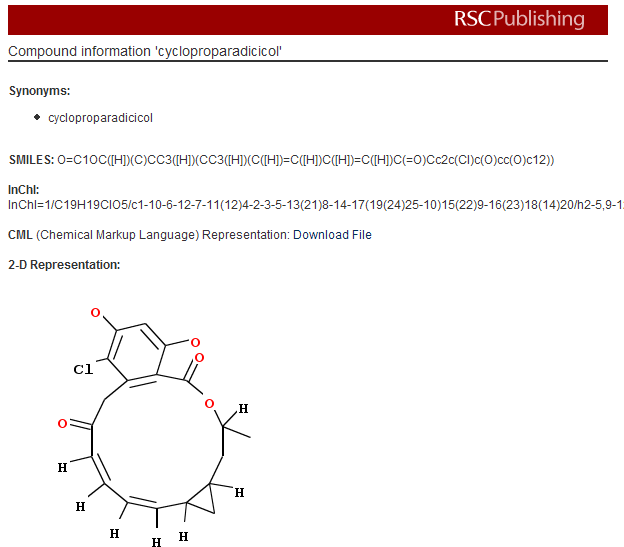

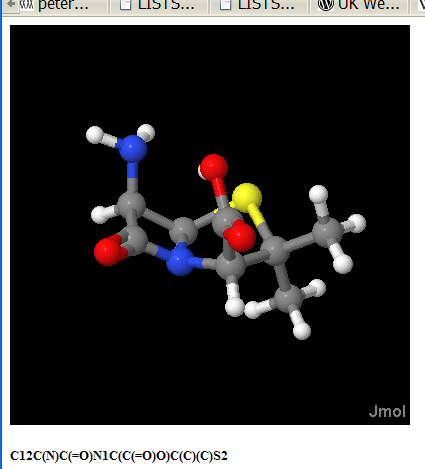
October 13th, 2007 at 8:31 pm ePeter, I will not announce the publisher at present because I made a commitment to not do so until we had a mutually agreeable blog posting for our users and accurately representing the conversation and agreements between us. I have an urge to co-exist in the world with publishers since they put a lot of value into the world. With the changes going on in Open Access figuring out how to co-exist is very necessary. I hope we can get the information out shortly. It is possible we have mis-stepped but more likely that there is a policy issue with spidering policy that needs addressing by the publisher.