NOTE: even if you are not a chemist, this should be worth reading…
Rich Apodaca evangelises the value of JRuby as a simple way of glueing together the Java offerings in our Open toolkit. Here he shows how easy it is to use Opsin for converting chemical names to structures. It’s worth reading in full if you are a chemical hacker (but I’ll omit the code here…) . After that I make some comments on OPSIN. Note that OPSIN is commonly distributed with the OSCAR3 package and the names are sometimes confused; OPSIN is a standalone tool for name2structure.
Recent articles have discussed the use of JRuby for cheminformatics. We’ve seen how to parse SMILES strings, and read or write InChIs. In this article, we’ll see how easy it is to parse IUPAC nomenclature from JRuby using Peter Corbett’s OPSIN library.Installation
After installing JRuby, simply download the OPSIN jarfile and copy it to your JRuby lib directory. You’re done.
A Simple Library
We can write a simple library to convert an IUPAC name into a CML document:
[details snipped; seeJRuby for Cheminformatics: Parsing IUPAC Nomenclature with OPSIN]
This simple Ruby library has parsed the name ‘4-iodobenzoic acid’ and has returned a string containing the CML representation for the molecule. If we had wanted the read_name method to return a traversable XML object model, we could have enabled that as well.Conclusions
One of the objections raised whenever the issue of “new” programming languages comes up, regardless of their merit, is the age-old refrain “Yeah, but where’s the software?” With JRuby, we bypass this question altogether. We can leverage the full scope of the massive Java development effort over the last ten years, which includes several excellent cheminformatics libraries. With virtually no effort, we have a working cheminformatics platform based on a widely-used, versatile and dynamic object-oriented scripting language. Future articles will discuss extensions to this platform and some applications.
PMR: Absolutely true. I don’t do a lot of glueware so I don’t use these languages, but my colleagues do.
OPSIN… I am sure Peter Corbett will add more, but the roots of OPSIN are about 5 years ago when we started the OSCAR work. OSCAR started as the Experimental Data Checker sponsored by the Royal Society of Chemistry, and after the first year it became clear that much of the data checking could not happen unless we knew the chemical structure. You can’t check that a compound has the right NMR spectrum if you don’t know how the atoms are connected.
Current chemical publishing and publishers destroy huge amounts of useful chemical information.
(Reread this sentence if you don’t believe it).
If you doubt it, visit the RSC’s Project Prospect (which is based on our collaboration over OSCAR). Find an example paper. (Most of the papers are closed, but there are a few open examples (e.g.) so you can see how it works). By default you will see a normal HTML page. However if you switch on the prospect enhancements, and select “Show Compounds” you should see the compounds highlighted
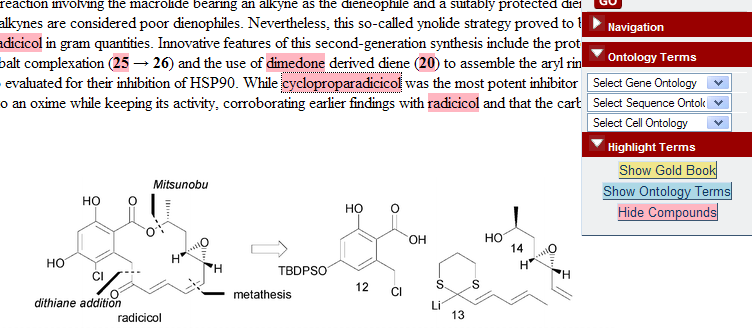
PMR: now I haven’t a clue what cycloproparadicicol is. I don’t even know how to pronounce it as it’s a made-up name. It’s not very interesting, in fact, as there are only 4 references in Pubmed and all to synthetic studies. It’s not interesting enough to be in Pubchem.
What Project Prospect does is to add this information in the paper. Click the link and you get a box like this which tells you what the molecule actually is:
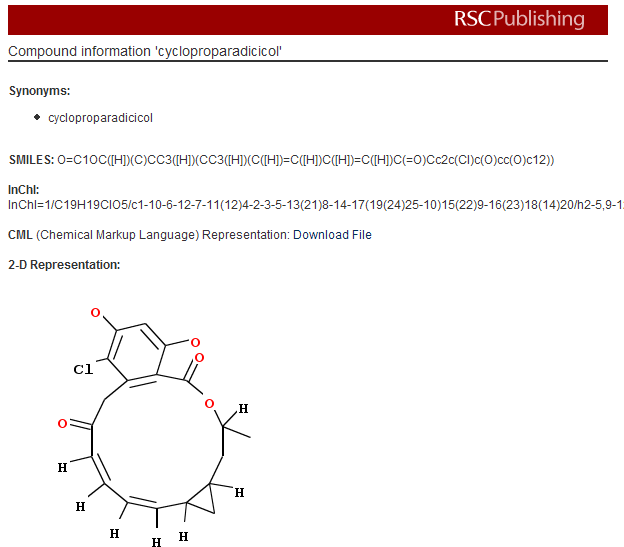
PMR: The irony is that the author knew all this when the paper was submitted but that there is no way of conveying it to the publisher. Publishers have no mechanism for authors to submit semantic info. And apart from these first starts at the RSC none of them have shown the slightest public interest during the last 10 years since e-publications became routine.
A simple analogy. When a food manufacturer gets wheat they throw out all the brown stuff that is good for you are produce white bread. It’s worse for you, the eater, but easier for them to produce. PDF is similar to white bread. Pretty but horrible and bad for your semantics. So Prospect is adding back the goodness that the publishing process takes out.
The problem is therefore that when robots read papers they don’t know the connection tables (e.g. the “SMILES” or “InChI” representation of the molecules above). So the only way to get the structure (in most cases) is to try to interpret the name. “4-iodo-benzoic-acid” is relatively easy to work out; cycloproparadicicol is impossible without a lookup table and there isn’t one. (I expect CAS has the structure but it will cost you 6 USD for each lookup).
If authors added InChIs and SMILES to their papers, much of the problems would be solved. The world would save thousands of person-years work. So why, over 10 years, haven’t we started doing it?
Mainly cock-up – I have only seen 1-2 publishers who have the slightest idea what is going on in Web 2.0 for example. Nature is one. But a small amount of deliberate opposition. I’ll let you see where that idea leads.
Until a brighter day dawns, with semantic papers, we’ll need InChI to get around the appaling lack of products that the current chemical publishers provide.
Pingback: Unilever Centre for Molecular Informatics, Cambridge - petermr’s blog » Blog Archive » OPSIN/OSCAR: you + us = we; please help
Pingback: ChemSpider Blog » Blog Archive » Name to Structure Conversion - and What One Little Patent Might Do…
Thanks for the description of Project Prospect, Peter. There are more free to read examples here.
Sitting here as a publisher, we don’t have half the power you suggest – we have to satisfy authors (we want the best) as well as readers (to give them the best service), and making submission as easy as possible is an absolute requirement. Demanding InChIs from authors isn’t a realistic option yet – we can show the advantages of this information via the enhanced HTML and work towards it, but compulsion’s an attractive but ultimately futile option. The more you push, the worse data you’ll get.
The publishers aren’t the problem – it’s because the possibilities of processing and reusing this information have only comparatively recently been apparent, and frankly because most people want to read the text and look at the pictures. As authors and readers are encouraged to look beyond print/PDF it’ll happen, but keeping the data within the publishing process is a community issue rather than a publisher one. We’d love it of course!
(3) Thanks Richard.
I don’t disagree with your analysis though very few publishers are even making noises about trying to move forward. It won’t stop me pushing.