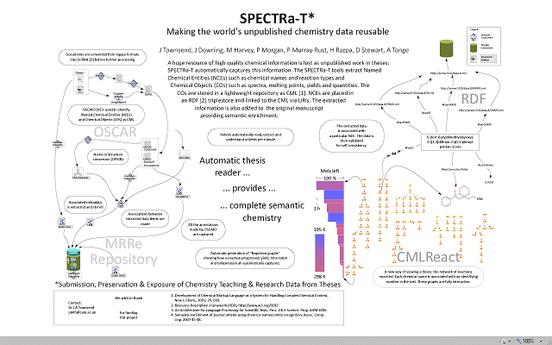
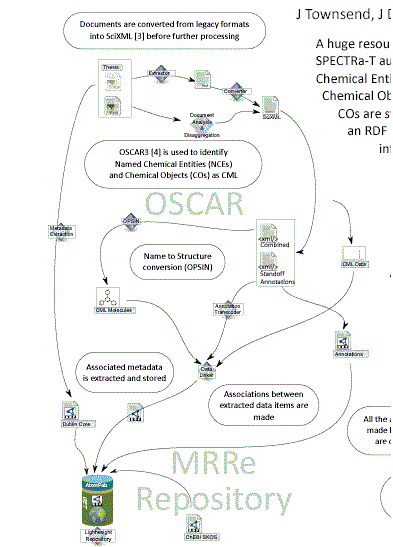
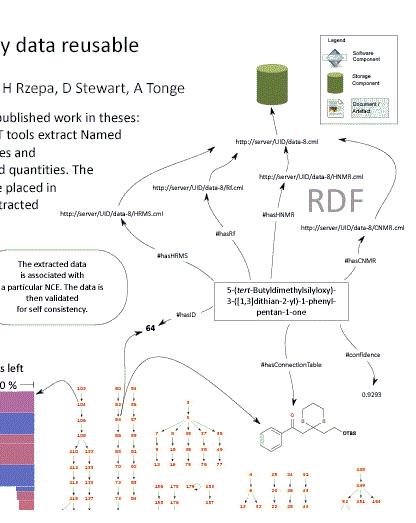
There is a major effort worldwide to capture student theses as born-digital objects. Much of the effort to date has concentrated on PDF documents as a proxy for the printed version, but PDF cannot achieve the full potential value of born-digital theses. Publication of properly structured theses can allow rapid exploration of material, for example down to individual diagrams or the sub-subsection level. Individual citations can be extracted automatically. Supporting data can be associated with the thesis directly rather than reduced to tabular or graphical form in appendices. Future readers will want not only the document level descriptive metedata (author, institution, title, keywords, dates) but also access to subcomponents (data, tables, references, diagrams, etc.). For example a reader might wish to survey all the analytical instrumentation used within the last year and therefore only need access to the “Materials and Methods” sections, but to wish this for every thesis. Similarly a bibliographer might wish to analyse communities of practice from citations. What proportion of references are identical to those in earlier theses from the same institution? Longitudinal studies may be possible – retrieve all images of protein gels over the last 10 years and see whether the quality has increased.
Current package-based approaches to the transfer and publication of complex digital objects result in problems of balkanisation due to the multiplicity of available packaging standards that complicate interoperability between systems and the discovery and reuse of complex object components. They usually use a “pass-by-value” paradigm of data transfer, which creates additional complexities in data duplication, trust / access control and revisioning. The ORE “pass-by-reference” approach allows a much more flexible, disaggregated approach to complex object description, access and transfer.
There is a growing realisation in the repository community that the web is not only an essential part of content delivery, but also an underused and undervalued architectural substrate for the repository ecosystem that offers easier interoperability between systems within our domain, and with the web community at large. ORE works within the constraints of the web architecture, therefore it will be possible to combine it with other web standards (e.g. Atom Publishing Protocol, SWORD, XACML) to integrate large scale infrastructures.
Project Aims
The general aims of the project are as follows:
Test the applicability of the ORE standard in a realistic scholarly setting – thesis description, submission and publication.
Demonstrate the advantages of the ORE approach in complex object publication, by combining it with existing web-standards compliant technologies.
Provide examples to fully exercise the ORE specification in order to provide validation and future direction.
Project Description
The experimentation in TheOREM will contain two main strands.
We will create a small corpus of ideal born-digital theses based on real theses (see this page) and describe these as completely as possible using ORE (see this page).
We will define a realistic scholarly scenario in which such theses might be handled, and implement demonstrators for each component system in the scenario in order to show the capabilities and limitations of ORE (see scenario-creation for details of the scenario notes on implementation).
We will investigate how ORE and modern web technologies can help with Embargo.
Project Discussion
Click here to view the wiki pages detailing current discussion about the TheOREM project. The ICE team’s trac also has project documentation and progress.
In it you will find the complete scenario for the submission of a thesis. I will hope to take the ETD2009 audience through it. I think the pictures are beautiful.
Like the current submission scenario, the updated version consists of 6 parties, which are:
The student doing the degree.
The students supervisor.
The students viva examiners.
A departmental thesis management system.
The Graduate Studies Office.
An Institutional Repository.
These are shown in the diagram below. The pages linked further down the page, contain similar images to describe the interactions during that part of the submission process. The position of the parties are kept constant, though for each component all parties may not be included in each image.

The updated submission process itself can be broken down into 6 sections:
Authoring of the thesis and embargo.
Initial submission.
Arrange the examiners and send them the thesis.
Doing the viva examination, fixing any corrections and resubmitting.
Approval of the final submission of the thesis and award of the degree.
Management of embargoed data.