[Readable by non-chemists].
Chembark is a well-known and highly respected chemical blogger. He and others keep the community on its toes in many ways, but often in revealing sloppy or even fraudulent practices. His latest post reveals unacceptable practice in publishing chemical experiments http://blog.chembark.com/2013/08/06/a-disturbing-note-in-a-recent-si-file/ “A disturbing Note in a Recent S[upplemental] I[nformation] File”. Supplemental (or Supporting) Information is the data (or usually a small part of the data) collected in the experiment and which is used to judge whether the authors have made the compound(s) they say they have. Chemists make a compound, purify it, collect spectral data and also analyse the elements in it (Elemental Analysis – how much Carbon, Nitrogen, Hydrogen – we can’t do Oxygen). Chembark’s commenters are split as to whether EA is useful but all agree it is required. (I am in favour of it – it may show that the wrong metal has been used – spectra won’t do this).
Here’s the paragraph from the SI. Even if you don’t understand chemistry you can see it’s the name of a compound (13), a recipe and some data.
This is very formulaic, indeed so formulaic that machines can understand it and 10 years ago 3 summer students and I developed a tool (OSCAR) which could read the numbers. It’s available as FLOSS and available to any publisher who wants to check data. I doubt that many do. (By contrast the crystallographers must submit and validate data before it is published. The current problem, which we’ll get to could never have happened in crystallography.
Here’s the problem – it occurs in the same file (http://pubs.acs.org/doi/suppl/10.1021/om4000067/suppl_file/om4000067_si_002.pdf ): Read it to the end
[Pt2(II)((M,SS,SS)-p-tolyl-binaso)2(μ-Cl)2][BF4]2 (14): A vial was charged with 100mg
(0.126 mmol) 5a and 24mg (0.126 mmol) AgBF4. 2ml CH2Cl2 was added, the vial was
covered and the reaction was left stirring in the dark for 2 hours. After this time, the
reaction was filtered over celite to remove AgCl. Solvent was then removed to leave a
yellow residue in the vial, the remaining clear, yellow solution was concentrated to a
volume of about 1ml, and diethyl ether was added in a dropwise manner to the stirred
solution to precipitate a yellow solid. The vial was centrifuged so the supernatant solvent
could be decanted off by Pasteur pipette. The yellow solid was washed twice more with
ether and the dried completely under high vacuum to give 99mg (93% yield) of product.
Emma, please insert NMR data here! where are they? and for this compound, just make
up an elemental analysis…
So this compound does NOT have any data. You’ve probably spotted the problem, but let’s show that a machine can also spot it. We run it through ChemicalTagger (developed by Lezan Hawizy in our group). ChemicalTagger (you can try this at home) reads a chunk of text and understands the semantic of chemical recipes. Everything coloured makes chemical sense included interpreting compounds and quantities:
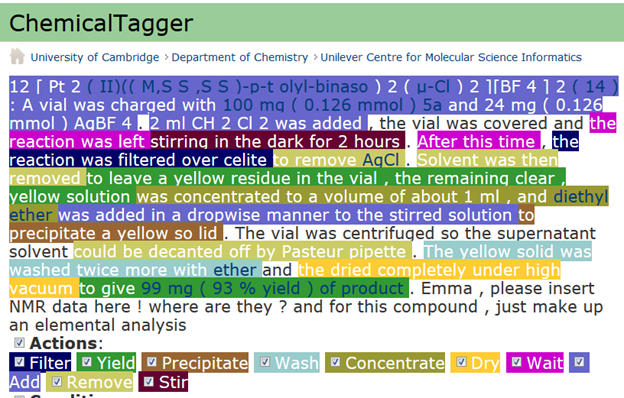
The last sentence leaps out as non-chemical! Now I’m not judging the authors (Chembark and his readers will investigate). But it should never have happened. If the file had been a data file, instead of a PDF software would have detected this before it was submitted. This is not a new idea, I have been beating my head against chemists and their publishers for ten years. They are completely uninterested in publishing semantic data.
It’s actually easier to publish semantic data than this PDF. If they published properly errors of this sort could not occur – a data referee would have picked this up. The spectra are all digital – they could be zipped up. But instead they are printed out and copied (see the splodges? That’s not experimental noise, probably coffee or JPEG destruction):
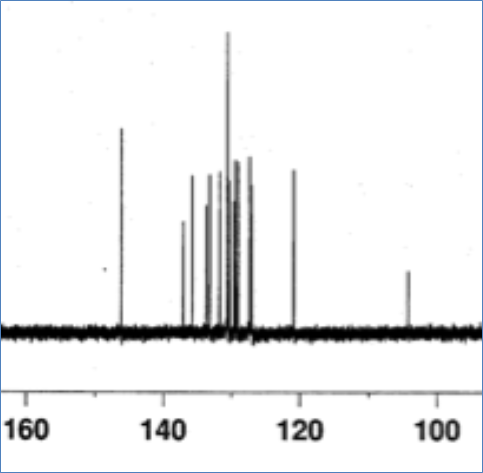
So until chemistry wakes up and manages its data they will consistently get chemistry whose supporting data is questionable. And that means questionable science.
I fully support publishing semantic data for a multitude of reasons, but one of the top ones is what you pointed out – JPEG destruction of spectra caused me nightmares when writing up papers and my dissertation. If the spectra wasn’t hi-res, it was kicked back to me, even when I worked up the data in a program like MNova or iNMR and copied it directly into the document! Honestly, just zipping up the data would be so much easier and so much nicer. I spent more time trying to get the damned spectra hi-res than I did actually writing my manuscripts!
Thanks so much Dave,
I hope to highlight your reply later.
You have to feel for the poor authors though, don’t you!
Years ago, I picked up an error like this late in production of one of my papers. When I am using latex, I *always* markup comments; this makes is pretty hard to leave in the final version. So, in effect, (some) authors already use a form of semantic markup. Then it gets thrown away.
It would be much easier to do this sort of thing for ourselves, if there weren’t any publishers in the first place. Now there is an innovative idea.
Thanks
RSC are starting with mol and jdx spectra as ESI, e.g.
http://pubs.rsc.org/en/Content/ArticleLanding/2013/CC/C3CC43488E
Thanks Richard,
Is this discretionary or mandatory? If the former then it is only enthusiasts who will use it (ACS has allowed many formats for many years and they have very little takeup). If mandatory I will applaud it and probably harvest the data as we do for Crystaleye.
Discretionary for now. Community acceptance needs to come before mandate, and to help that we’ll be building reuse cases to show the value.
Thanks,
I will see if we can rerun the Crystaleye software to extract JDXs, etc. I assume that all formats will be ASCII