Jenny Molloy has invited me to introduce a session in Oxford this evening (Monthly meetings held at Oxford e-Research Centre, 7 Keble Road,
19:00-20:30 – See more at: http://science.okfn.org/community/local-groups/oxford-open-science/#sthash.h1XPy1Pm.dpuf and http://science.okfn.org/community/local-groups/oxford-open-science/ ) on Content Mining. It will be very informal – anyone can come and play.
The basis theme will be that:
- Content mining is now routinely possible
- YOU can do it
- Your involvement will be a massive help
The idea is to build a community and collect/create community tools. There’s been a massive step forward with Jailbreaking the PDF (http://www.duraspace.org/jailbreaking-pdf-hackathon ) and I’ve blogged some of this (/pmr/2013/05/28/jailbreaking-the-pdf-a-wonderful-hackathon-and-a-community-leap-forward-for-freedom-1/ ). We also had a tremendous hackday in London (http://hack4ac.com/ ) (see Ross Mounce’s blog http://rossmounce.co.uk/2013/07/09/hack4ac-recap/ ). This was run by new-generation publishers (Ian Mulvany (PLoS), Jason Hoyt (PeerJ)) and looked at how hacking can change scholarly publishing. We came up with several ideas (I’ll blog my own soon) and Ross proposed figures2Data – what can we extract from the *figures* in the literature (not just the text).
We got a critical mass of 4-5 people and a great reservoir of knowledge and ideas. We made fantatstic progress. This is a very difficult subject and I assumed we wouldn’t manage much. However we found communal resources showing it can be done relatively simply using classical Computer Vision (Image analysis) such as Hough transforms and character recognition. I’d known that I’d have to hack the latter some time and was dreading it, but we found that Tesseract (http://en.wikipedia.org/wiki/Tesseract_%28software%29 ) would provide a huge amount out-of-the-box. This is a great example of stepping back from the problem and letting in fresh light. I’m now pretty confident that we can manage to hack a wide range of scientific diagrams. Here’s a diagram
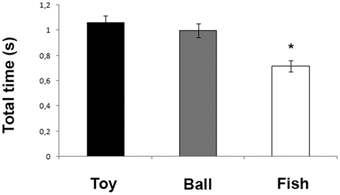
And here is what Thomas managed to extract (in an hour or two):
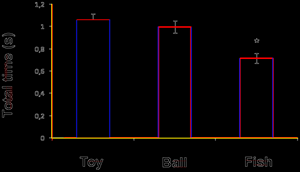
The OpenCV http://en.wikipedia.org/wiki/OpenCV suite is tremendous and so powerful that navigation is a problem but Thomas has found all the key components. I am confident that Tesseract will recognize the characters and so we are well on the way to extracting all the information from this.
As readers know, Ross and I are working on phylogenetic trees (hypothesising the formation of species). The great thing about this subject is that anyone interested in science should be able to understand the basic concepts. It’s particularly useful for conservationists, biodiversity etc. So we are seeing what can be extracted from papers on this subject. I’m told there may be some people tonight specifically interested in this. This is a great area in which to start practical open-science.
The main problem, alas, is that most publishers have put in place legal restrictions to stop us doing this. There is no scientific reason for this. It’s to “protect their commercial interests”. So Jenny, Diane Cabell and I reviewed this for the Open Fellowship Academy http://www.openforumacademy.org/library/ofa-fellows-reference-library p57: There we put forward a manifesto for content-mining under the mantra
The right to read is the right to mine.
Simply – if you have paid to be able to read science your machines should also have the right to read it.
But no, the publishers are fighting this and trying to licence the “privilege”. It’s being debated in Europe (Licences 4 Europe) and Ross presented a superb set of slides (http://www.slideshare.net/rossmounce/content-mining) and this will give you a good indication of the technology and the restrictive practices.
So I’m hoping that tonight will see an expansion of our content-mining community!