#ami2 has been working on converting PDFs to scientific XML and we have made significant progress. Today we converted a page of PDF to chunks of SVG and then aggregated these to XHTML and non-textual chunks. We’ll show some of the technology below. Ross Mounce @rmounce and I will be demo’ing at Beyond the PDF2 next week at Amsterdam. In brief
AMI2 is an open collaborative platform which can take existing scientific PDFs and turn them into semantic science
Everyone is invited to join in. The abstract infrastructure is well advanced. We’ll soon be re-adding vector-graphics-driven graphs and trees these should be available for demo next week. Possibly some simple tables. Of course there will be a long tail of minor problems but for many papers that won’t matter At this stage, therefore we’d like:
- Anyone who is interested in having PDFs converted to bring some and we’ll see how we get on. The things that cause problem are non-standard Fonts and bitmaps. Probably takes 15-30 secs to convert a paper so bring some on a USB stick. Theses should also work.
- Volunteers to join the effort. You don’t have to be a coder, but you need a hacker mentality. You can help by adding Font info, checking the output visually, creating a corpus, etc.
- Hackers interested in tackling general graphics artefacts. The most common are graphs, barcharts, block diagrams, flowcharts,
- Domain experts (chemistry, phylogenetic trees, species, maps, sequences …)
- Hackers for math equations
- Analysis of bit-mapped graphics (harder, but tractable)
We’re already building up some fantastic Open viewers which we’ll show at Amsterdam. So far we have:
- Visomics (from Kitware) which allows interactive browsing of phylo trees linked to the Open Web (e.g. Wikipedia species)
- JSpecview – an interactive spectrum viewer allowing peak picking comparison, integration, etc.
- JChempaint – 2D molecules display and editing
The great thing is that as a community we can choose those areas which are most important to work on. We then have an area of tools which make the scientific literature more valuable than the dead PDFs.
Here’s an example of a mixed page of text and graphics. From an openly visible artcil;e in the CSIRO Australian J Chem:
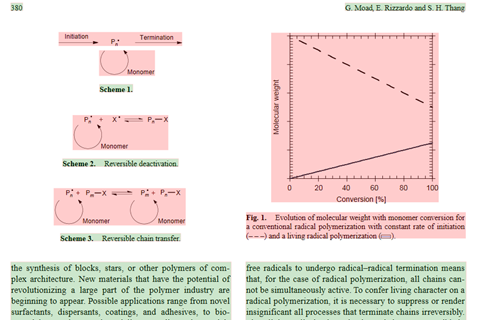
SVG2XML has identified the chunks in the page, and their role – in this case text (green) and mixed text and graphics (red). The text is then turned into XHTML and here’s the result. Notice that it’s free flowing and can warp, be scaled, etc. AMI2 manages fonts (and there are some seriously non-standard ones)and sub/superscripts. It can be edited, re-used reformatted, turned back into PDF – whatever you want.
[NOTE. There will be a wail – why don’t you use the publishers’ XML. Simple… It’s behind a paywall and not re-usable legally. But that’s no longer a problem. We can probably omit the publishers’ XML (BTW NEVER let a publisher charge you extra for XML)
So here’s the XHTML. It’s totally readable by machines and humans. And before the type-setters tell us this doesn’t look as good as their creation, this is hot off the press and there are lots of automatic processes we can apply to make it quite acceptable.
Interesting. Is there a homepage for AMI2 out there, to learn more about it?
http://bitbucket.org/petermr/pdf2svg
http://bitbucket.org/petermr/svg2xml-dev
From OKF list
I and collaborators are very actively working on this and making all
> resources available under OKD-compliant terms. The project is code-named
> “AMI2” and is initially targeted at scientific and technical publications.
>
> We use the excellent Apache PDFBox http://pdfbox.apache.org/ to extract
> the contents of the PDF and are layering tools on top of this. The end
> product – RSN! – will be XHTML with domain-specific inserts for math,
> chemistry, graphs and tables. The vision is to read millions of papers each
> year and extract data automatically.
>
> We would very much welcome a community effort to help with things like
> character encodings, sample material , knowledge of fonts etc.
> alpha-testing, etc. The work is blogged under #ami2 on my blog – see
> /pmr/2012/10/20/opencontentmining-the-ami2-project-for-understanding-the-scientific-literature-and-why-i-love-pdf/.
>
> Be aware that “PDF” is not a simple concept. It can include:
> * photographs of paper artifacts (challenging)
> * scanned text (requires OCR)
> * text with characters (common in modern PDF publications)
> * text as vector graphics (i.e. no font info)
> * bitmaps of tables, graphs, etc.
> * vector graphics of graphs etc.
>
> Where the document has vector graphics (which come from EPS files, PPT,
> etc.) or characters the ease and quality of extraction is much higher than
> for bitmaps. So whether any particular instance is feasible depends on
> details.
>
> Ross Mounce (OKF Panton fellow) and I are working very actively on this
> area.****
>
> ** **
>
> For the next School of Data tutorial I would like to cover Data extraction
> from PDFs (and text) as well as OCR.
>
> Does anyone here have experience with Tools that do not require coding
> skills to extract data and text from PDFs?****
>
>
> It’s my intention to make this an automatic process – we have a little way
> to go but it’s months not years.
>
> We’d be very interested in any who likes hacking this sort of thing. ****
>