Ross Mounce and I are now geared up for content mining of the bioscience literature, and I’ll be giving you some idea of the technology. There are many publishers who either won’t let us content mine (ACS, RSC) or who are “very helpful” but either don’t reply or mumble (e.g. Universal Access). So we are starting with good honest CC-BY as promoted by Gulliver Turtle and friends at BMC. So all our illustrations will come from BMC. (Why not PLoS? These are technical, not political restrictions we’ll analyse later. We’d like to do PLoS).
So I shall give simple discussions on archetypal content mining and today we will start with the tree. (or Larch if you follow the Pythons).
Here’s a tree: from

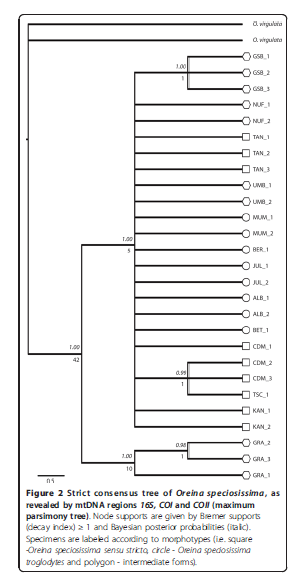
And here’s the bottom bit magnified:
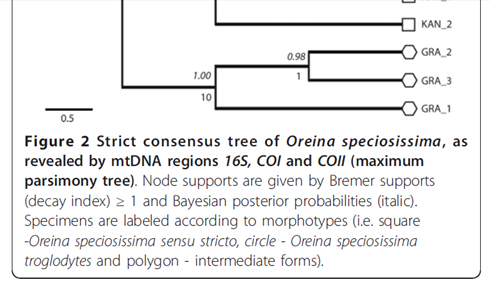
Questions:
- What makes a tree?
- Can machines determine whether it’s a tree (a) from the tree (b) from the caption (which we have as running UTF-8 text?
- Why is this particular example not well suited for content mining?
- Are there other disciplines than bioscience where trees (in the abstract) occur?