#oscar4
Follow http://www-pmr.ch.cam.ac.uk/wiki/OSCAR4_Launch for all news and URLs (live stream). Hashtag #oscar4.
To introduce the launch of OSCAR4 I will give a short timeline and hopefully credit those who have made contributions.
When I joined the Unilever centre I had the vision that the whole of chemical information could be used as a basis for intelligent software agents – a limited form of “artificial intelligence”. By collecting the world’s information, machines would be able to make comparisons, find trends, lookup information, etc. We would use a mixture of algorithms, brute force, canned knowledge, machine learning, tree-pruning, etc.
The knowledge would come from 2 main sources:
- Data files converted into semantic form
- Extraction of information from “written” discourse. This post deals with the latter – theses, articles, books, catalogues, patents, etc.
I had started this is ca 1997 when working with the WHO on markup of diseases and applying simple regular expressions. (Like many people I thought regexes would solve everything – they don’t!).
In 2002 we started a series of summer projects, which have had support from the RSC, Nature, and the IUCr among others. We started on the (very formulaic) language used for chemical synthesis to build a tool for:
- Checking the validity of the information reported in the synthesis
- Extracting and systematising the information.
This was started by Joe Townsend and Fraser Norton and at the end of the summer they had a working applet.
2003
Sam Adams and Chris Waudby joined us and turned the applet into an application (below); Vanessa de Sousa created “Nessie”, a tool which linked names from documents to collections of names (such as collections of catalogues (at that stage these collections were quite rare).
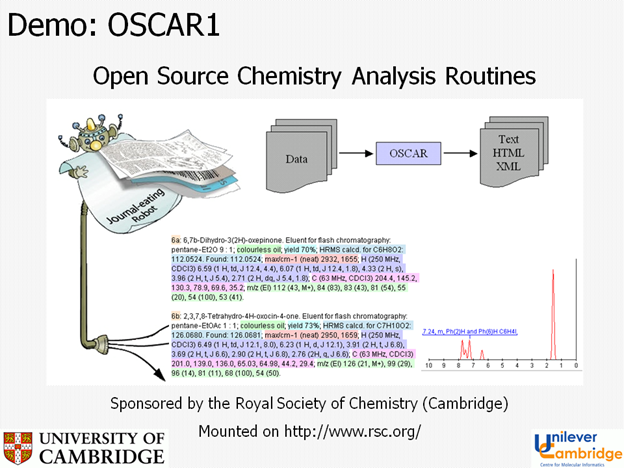

Here is OSCAR-the-journal-eating-robot in 2004 eating a bit of journal, analysing it, validating it and transforming the data into CML (e.g. a spectrum).
By 2004 OSCAR was in wide use on the RSC site (http://www.rsc.org/Publishing/Journals/guidelines/AuthorGuidelines/AuthoringTools/ExperimentalDataChecker/index.asp ) and could be downloaded for local work. During this period the variants of OSCAR were lossely named as OSCAR, OSCAR-DATA, OSCAR1 and OSCAR2. I pay tribute to the main authors (Joe Townsend, Chris Waudby and Sam Adams) who have produced a complex piece of software that has run without maintenance or complaint for 7 years – probably a record in chemistry. At this time Chris developed an n-gram approach to identifyig chemical names (e.g. finding strings such as “xyl” and “ybd” which are very diagnostic for chemistry).
By 2005 OSCAR’s fame had spread and (with Ann Copestake and Simone Teufel) we won an EPSRC grant (“Sciborg“) to develop natural language processing in chemistry and science (http://www.cl.cam.ac.uk/research/nl/sciborg/www/ ). We partnered with RSC, IUCr and Nature PG all of whom collaborated by making corpora available. By that time we had learnt that the problem of understanding language in general was very difficult indeed. However chemistry has many restricted domains and that gave a spectrum of challenges. Part of SciBorg was, therefore, to develop chemical entity recognition (i.e. recognising chemical names and terms in text). Simple lookup only solves part of this as (a) there are many names for the same compound (b) hypothetical or recently discovered/synthesised compounds are not in dictionaries. This work was largely carried out by Peter Corbett (with help from the RSC) and the tool became known as OSCAR3.
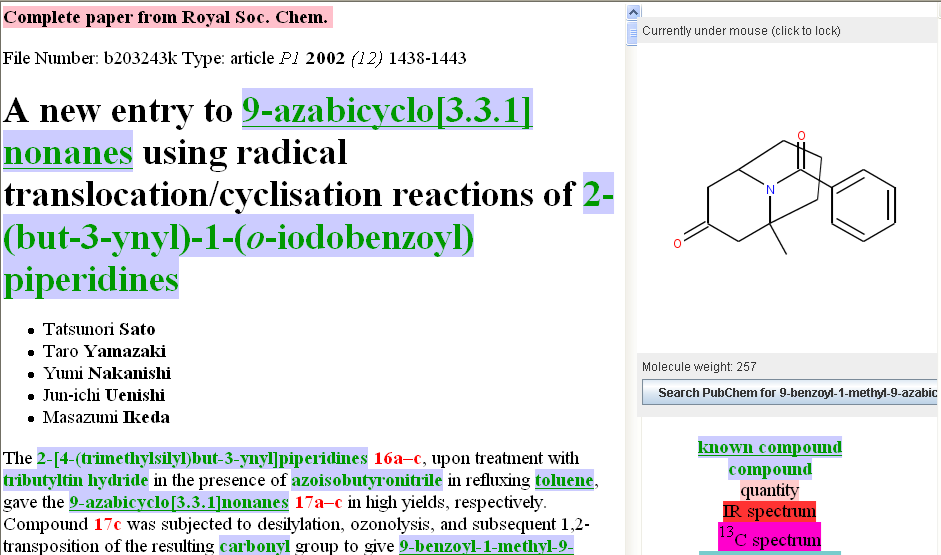
Entity recognition is not an exact science and several methods need to be combined: lookup, lexical form (does the word look “chemical”), name2Structure (can we translate the name to a chemical?), context (does it make sense in the sentence?) and machine learning (training OSCAR with documents which have been annotated by humans). There are many combinations and Peter used those which were most relevant in 2005. OSCAR rapidly became known as a high-quality tool.
This required large amounts of careful, tedious work. A corpus had to be selected and then annotated by humans. Strict guidelines were prepared; here’s an example from Peter Corbett and Colin Batchelor (RSC). What is a compound? There are some 30 pages like this.
. 
The humans agreed at about 90% precision/recall, setting the limit for machine annotation (OSCAR is about 85%). OSCAR can do more than just chemical compounds and here is a typical result:
In 2008 Daniel Lowe took up the name2Structure challenge and has developed OPSIN to be the world’s most accurate name2structure translator, with a wide range of vocabularies. OPSIN was bundled as part of OSCAR3 which also acquired many other features (web server, chemical search, annotation, etc.). We were joined by Lezan Hawizy who initiated Unilever-funded work on extracting polymer innformation from the literature. Joe Townsend worked on the JISC SPECTRaT project looking at extracting information from theses. In 2009 we won a JISC grant (Cheta) to see whether humans or machines were better at marking upn chemistry in text and work proceeded jointly at Cambridge (Lezan Hawizy) and the National Centre for Text Mining (NaCTeM, Manchester).
In late 2009 OMII contracted to start the refactoring of OSCAR and in 2010 we won an EPSRC grant to continue this work in conjunction with Christoph Steinbeck (ChEBI at EBI), and Dietrich Rebholz. The hackforce included David Jessop, Lezan Hawizy and Egon Willighagen resulting in OSCAR4.
OSCAR is widely used but much of our feedback is anecdotal. It’s been used to annotate PubMed (20 million abstracts) and we have used it to analyse > 100, 000 patents from the EPO (“the green chain reaction”; example: http://greenchain.ch.cam.ac.uk/patents/results/2009/solventFrequency.htm ). In 2010 we were funded by EPSRC “Pathways to Impact” to extend OSCAR to Atmospheric chemistry (Hannah Barjat and Glenn Carver).
Besides the people mentioned above thanks to Jonathan Goodman, Bobby Glen, Nico Adams, Jim Downing.
Summary:
Thanks to our funders and supporters: EPSRC, DTI, Unilever, RSC, Nature, IUCr, JISC.
OSCAR4 now has a robust architecture and can be extended for work in a variety of disciplines. It interfaces well with other Open Source tools (e.g. from the Blue Obelisk) and can be customised as a component (e.g. in Bioclipse) or using OS tools and its own components.The main challenges are:
- The commonest form of scientific document, PDF, is very badly suited to any information extraction.
- Almost all publishers forbid the routine extraction of information from their publications.