This is a post in a series outlining Open Semantic Data in Science at BioIT Boston – see (BioIT in Boston: What is Open? ).
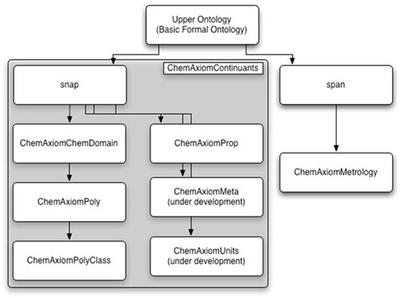
To build a complete semantic framework we need formal systems to express our concepts, normally markup languages or ontologies. In Cambridge we use both, particularly Chemical Markup Language and ChemAxiom (Nico Adams). Let’s start with ChemAxiom which Nico has blogged. His diagram gives a good overview:
Since Nico leads our Polymer Informatics effort there is a special concentration on polymers but the framework is very general and can be used for mainstream chemistry. As shown ChemAxiom emphasize substances and their properties. Because of our backgrounds as (in part) physical chemists there is emphasis on methods or measurement (metrology) and scientific units of measurement.
The ontology is descended from the Basic Formal Ontology which is an Upper Ontology. This has abstracted concepts which are common to many different ontologies – science, commerce, literature, etc. From WP:
The BFO or Basic Formal Ontology framework developed by Barry Smith and his associates consists in a series of sub-ontologies at different levels of granularity. The ontologies are divided into two varieties: SNAP (or snapshot) ontologies, comprehending continuant entities such as three-dimensional enduring objects, and SPAN ontologies, comprehending processes conceived as extended through (or as spanning) time. BFO thus incorporates both three-dimensionalist and four-dimensionalist perspectives on reality within a single framework
In this a compound would be a continuant whereas a reaction as performed by a given person would be an occurrent. Note that language often maps different concepts onto the same linguistic term:
“The reaction took place over 1 hour” (occurrent)
“The aldol reaction creates CC-bonds”
Is the latter use a continuant or occurrent? This is the sort of thing we discuss in the pub.
ChemAxiom is expressed in OWL2.0 and this brings considerable power of inference. For example if something is a quantity of type temperature the ontology can assert that Kelvin is a compatible unit while Kilogram is not. And this is only a simple example of the power of ChemAxiom.
In general we expect ChemAxiom to have a stable overall framework as expressed in the diagram but for the details to change considerably as the community debates (hopefully good-naturedly) over the lower level concepts. Nico is presenting this at the International Conference on Biomedical Ontology.
Chemical Markup Language uses a wider-ranging set of chemical and physical concepts which are derived from chemical communication between humans and machines in various degrees. We (Henry Rzepa and I) have used published articles, chemical software systems, chemical data and computational chemistry outputs to define a range of concepts which now seem to be fairly stable. That’s perhaps not too surprising as many of them are over 100 years old. The main concepts are:
- molecules, compounds and substances
- chemical reactions, synthesis and procedures
- crystallography and the chemical solid state
- chemical spectroscopy and analytical chemistry
- computational chemistry.
There are about 50 sub concepts, and all these are expressed as XML elements. Thus we can write:
<cml:property dictRef=”properties:meltingPoint”>
<cml:scalar dataType=”xsd:double” units=”units:kelvin”>450</cml:scalar>
</cml:property>
The general semantics are that we have a property, with a numeric value. The precise semantics are added by links to dictionaries (or ontologies such as ChemAxiom). With this framework it is possible to markup much of current chemical articles.
It’s also possible to markup much computational chemistry output and we’ve done this for many major codes such as Gaussian, Dalton, GULP, DL_POLY, MOPAC, GAMESS, etc. This has made it possible to chain together processes into a semantic workflow:
Here one program emits XML and feeds automatically into the input of the next. All output is semantic and can be stored in XML or RDF repositories such as our Lensfield which is being developed by Jim Downing and others
Which brings me nicely onto Data… in the next post
This Blog Post prepared with ICE 4.5.6 from USQ in Open Office

It strikes me that we can add CML markup to some of the properties we display on Wikipedia. I’m not sure if this would be useful for the community as a whole, but it may well be worth having a think about: it wouldn’t do any HARM, after all!