Totally Synthetic, Chemspider and I have been discussing the value of InChIs in blogs. TS’s blog is, of course Openly available under CC licence, and he is widely revered in the community for the beauty and acuuracy of his structural diagrams. This post is a slightly light-hearted voyage through what can be discovered with Toll-Access barriers in place. I leave readers to judge whether TSand Pubmed are up to the ease and value of the information from commercial providers.
I’m reading this from outside the University and I do not have a VPN. This is useful as it shows me what it’s like to be an information-impoverished reader. TS blogged today about Diazonamide A , a natural product which was billed as the next big breakthrough in cancer some years ago. (It has 4 reports in Pubmed about its biology, and 26 ones about the chemical synthesis. Taxol has 30,000). Anyway TS has taken the advice of the Blue Obelisk list and managed to put InChIs into his blog.
I’ll show his beautiful-as-always structure at the end, but meanwhile I wanted to see how easy it was to find the structure from freely accessible sites. This includes most abstracts (in science it seems to be almost universal to post abstracts in clear, so be grateful).
Wikipedia does not list it, but has the (intriguing and misleading) entry under “Trivial_name”:
For example, the most important structural feature of Diazonamide is that it’s a nonribosomal peptide, which is denoted by the suffix “amide“.
PMR: it might have started as a peptide but I don’t think many people would now call it that. (Unless there is another Diazonamide that I don’t know of).
So on to the latest synthesis (Magnus, Cheung, Goldberg, Russell, Turnbull and Lynch. JACS, 2007, ASAP. DOI: 10.1021/ja0744448.), remembering I can’t read the full text. The abstract is a superb illustration of hanging links (NullPointerExceptions in Java):
Abstract:
During the course of studies on the synthesis of diazonamide A 1, an unusual O-aryl into C-aryl rearrangement was discovered that allows partial control of the absolute stereochemistry of the C-10 quaternary stereogenic center. Treatment of 30 with TBAF/THF gave the O-tyrosine ethers 31 and 32 (1:1), which on heating each separately in chloroform at reflux rearranged to 33 and 34 in ratios of 84:16 and 56:44, respectively. This corresponds to a 70% yield of the correct C-10 stereoisomer 33 and a 30% yield of the wrong C-10 stereoisomer 34. Attempts to convert 34 into 33 by ipso-protonation and equilibration were unsuccessful. Confirmation of the stereochemical outcome of the rearrangement was obtained by converting 33 into 37, an advanced intermediate in the first synthesis of diazonamide A by Nicolaou et al. It was also found that the success of the above rearrangement is sensitive to the protecting group on both the tryptophan nitrogen atom and the tyrosine nitrogen atom.
PMR: What a splendid piece of non-communication! [My comments could apply to many publishers, not just ACS]. Without the full text (which, after considerable perusal will tell us what 1, 30, 31, 32, 33, 34 and 37 are) it’s almost meaningless. I am reminded of Alice’s comment on Jabberwocky:
“Somehow it seems to fill my head with ideas – only I don’t exactly know what they are! However, SOMEBODY killed SOMETHING: that’s clear, at any rate — ‘”
PMR: and the authors made something from something else…
So off to Pubchem. Many compounds made by synthetic chemists are no in Pubchem because they are of no interest, but Diazonamide is. It has a structural diagram [1]

PMR: Lovely. I think it’s correct, but it’s not exactly beautiful. like mathematical equations chemical strucures can be pretty or semantic. This is semantically correct and it’s probably pretty to jellyfish (this was a marine compound) but not to humans.
So on the InChI. Pubchem tells me that the compound has InChI:
InChI=1/C40H34Cl2N6O6/c1-15(2)27-37-46-29-32(54-37)40-20-9-5-8-19(18-7-6
-10-22-25(18)26(33(41)43-22)31-34(42)48-38(29)53-31)28(20)47-39(40)52-24
-12-11-17(13-21(24)40)14-23(35(50)45-27)44-36(51)30(49)16(3)4/h5-13,15-1
6,23,27,30,39,43,47,49H,14H2,1-4H3,(H,44,51)(H,45,50)/t23-,27-,30-,39-,4
0u/m0/s1/f/h44-45H
The problem is that this is not pretty for blogs as it runs over the line ends and spaces are a problem. So IUPAC are working out new approaches and some of these are discussed by the Blue Obelisk.
There is also a SMILES:
CC(C)C1C2=NC3=C(O2)C45C(NC6=C(C=CC=C64)C7=C8C(=CC=C7)NC(=C8C9=C(N=C3O9)C
l)Cl)OC2=C5C=C(CC(C(=O)N1)NC(=O)C(C(C)C)O)C=C2
which is a linear way of encoding the structure. Let;s go to the Daylight site (they invented SMILES) to see what it looks like:
I think it’s correct, and it’s certainly a lot better than the Pubchem offering but it’s not beauty – except for Shrek.
Let’s try Chemical Abstracts. It’s got every compound ever made. Maybe they will let me have a free go… (STNEasy) I find:
A free demo! Just what I wanted…
PMR: This is fine, and it points to the same abstract, but I can’t get at the structure. Let’s try CAS-Number lookup – it will tel me the number and the structure… and there is a free demo as well:
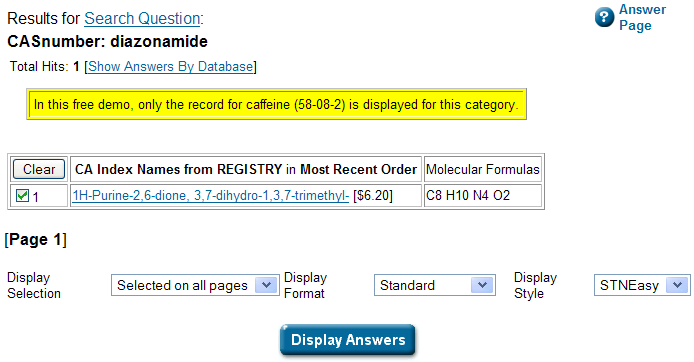
Oh dear… Yes, a free demo, but only if you are looking for caffeine. I get get all I want about caffeine from Wikipedia without paying 6.20 USD. Ah well,
So, off to chemspider which is free. The search for diazonamide A reveals:
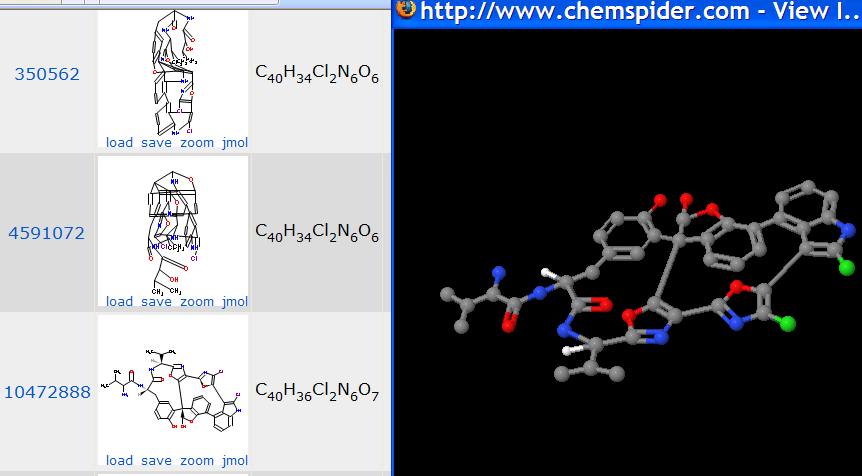
10472888 is shown at full size. (There are two more structures but both are equally unreadable). Note that the atom counts of the structures are inconsistent – the actual composition – I think – is that of 4591072. I try to zoom the formula and get a featureless gray square on both IE and Firefox. So I try Jmol (shown right). Now the molecules are three-dimensional but the coordinates in chemspider are those of the 2-D diagram. Personally I regard this as extremely misleading and would NEVER use Jmol for 2D diagrams, but I shan’t pursue this here.
So I still don’t know what the molecule is. Where else? Perhaps I can use some more abstracts…
And the fourth one on Pubmed hits gold. It’s from PNAS:

 and it’s FREE!!!!!
and it’s FREE!!!!!
so we find the structure:
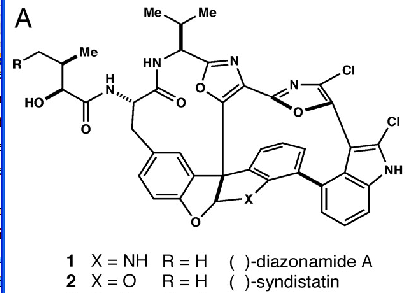
Truth at last. (For non-chemists the exact width of the lines matters, and the pixellation makes it very difficult to be sure. But I’m sure it’s correct.
And now what you have been waiting for – Totally Synthetic’s structure:

I think you’ll agree that the blogosphere is starting to emerge as a serious place to look for chemistry.
[1] pasted directly from the Pubchem site, suggesting we can create an image library for chemical structures
Pingback: ChemSpider Blog » Blog Archive » More Comments About Diazonamide A - other efforts to distinguish WHAT’S REAL?
Peter…I’ve spent some time tonight looking more deeply into the chemical structure details re Diazonamide A. Looks like we were thinking similarly as I made a post before I saw your blog posting. I then blogged a response to yours.
Your readers especially should be interested in the ChemRefer service I talk about to search publications online. Chemrefer is now associated with ChemSpider “by marriage”
http://www.chemspider.com/blog/?p=147
and
http://www.chemspider.com/blog/?p=153
This has given me an interesting opportunity to validate structures across databases. Seems we had the same intent. I’m asked Paul to send me the structure he drew to check stereo details.
For the record, you can compare with CDK’s SMILES to 2D at:
http://cheminfo.informatics.indiana.edu/~rguha/code/java/cdkws/cdkws.html#sdg
Interesting stuff. Let me add some additional points. One of the structures in PubChem comes from us (DTP/NCI). If you look at the compound record you get the mess you included above. If you navigate to the substance record (click on the CID, on that page look for the Substance: 1 link, when that hit comes up click on the SID, and on that page change the drop down choice for Compound Displayed from PubChem to deposited) you see a much more sensible 2D drawing. In a quick look, I think the difference between the two structures in PubChem is that one of the stereocenters in the NCI deposited structure is unspecified in the other structure. The NCI structure was submitted in 1997 by one of the authors of the original isolation paper. As the structure correction was published in 2001, it is almost certain that the NCI structure contains the original error. I say almost because we have no audit trail in our internal database for structures (at least not easily visible to me). A NSC has a structure, period. There is some possibility that the structure was fixed, but that just overwrites the previous structure. It just reinforces my view that it is extremely important to treat the structure of a substance as one more data point, subject to varied and possibly conflicting values.
This discussion demonstrates why I believe that “structure drawing” falls within the domain of materials than can be copyrighted. What is the difference in all of these representations (let’s agree not to worry about what might be a different stereochemistry at one center)? In the eye of the viewer some of these are “ugly” and some are more aesthetically pleasing. I might argue that Totally Synthetic’s representation is not only clear, it might even be called “pleasing”.
Since it is aesthetics (beauty, clarity, etc) that differentiates these drawings, the creator’s choices in how to display this molecule were critical creative acts. It seems to me that this defines work that should fall under copyright protection. The fact that all of these representations refer to the same underlying chemistry does not diminish in the least the creativity involved to create the last structure, and perhaps a total lack of creativity in producing the PubChem drawing.
So it seems to me that one should be careful in the re-use of structure drawings. To me these “drawings” are not data, while the connection table, for example, is data.
Steven