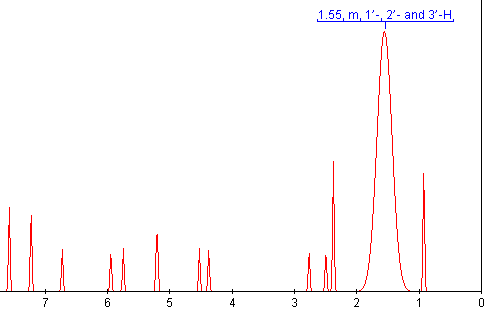
Sometime in the 1970’s the Amer. Chem. Soc. published a review of Computers in Chemistry (cannot remember date or title and I’ve lost my copy) and it has remained an inspiration ever since. In it was summarised the work of the Stanford (DENDRAL, CONGEN) and Harvard (LHASA) groups on the applications of artificial intelligence to chemistry (structure elucidation and organic synthesis). Both have heavy elements of problem-solving, coupled with pattern recognition. The systems effectively contained:
* a knowledge base of chemistry
* a set of heuristics (rules)
* formal deterministic procedures (e.g. tree searches).
The accomplishment was remarkable. The systems worked. They weren’t as good as a professional synthetic chemist, but in small areas they were better than me. It seemed obvious to me that with sufficient work on all components, but especially the knowledge base these systems would be able to do organic chemistry at the level of all except the best in the field. Certainly I expected that with the passage of 30 years the chemist/machine combination would be common. (Admittedly I sometimes believed too much hype about AI – now that I work in one branch (language processing) I know how difficult it is).
At the same time very similar work started to be done on chess. Again, when the first programs came out I could easily beat them (and I am a weak player). But gradually they improved and now they can beat essentially all humans.
It seemed to me that chemistry and chess would be quite similar. They are formal systems, too complex for brute force, and where a knowledgebase is essential. In chess all significant games have been captured in a database, and a large number of endgames have been exhaustively worked out. What is interesting is that the chess grandmasters have formed a symbiosis with computer programmers and machines and are still exploring what aspects machines can and cannot do. (I’m not an expert here and comments would be welcome).
By contrast there has been no significant work on chemistry and AI in, perhaps, 15 years. When I was in the pharma industry my boss used to speak of “another outbreak of Lhasa fever” (sic) – meaning that someone had suggested that machine synthesis should be explored. The Lhasa organisation has effectively stopped supplying synthesis methodology and turned to toxicology prediction (albeit it highly valuable).
So I feel a considerable feeling of sadness. I am sure that if synthetic chemists had embraced computers in the same way as chess players we would be sgnificantly better off. This is, of course, an act of faith but it’s borne out by the knowledge revolution taking place in many disciplines. The bioscientists are eagerly exploring the S/semantic W/web witn formal ontologies and reasonaing – another approach to “AI”.
I’ve just been at the UK eScience meeting (cyberinfrastructure) meeting for 3 days. (I’ll probably hark back in future posts). One keynote was given by Stephen Emmott (Director, Eur. Sci. Programme) Microsoft Research, Cambridge). Stephen talked about 2020 and gave a vision when computing could be based on biology – where molecular computers have already been injected into cells. Microsoft is hiring bioscientists who are also computer-able (i.e. they can make their ideas happen through code, rather than requiring comput/er/ational scientists to write the code for them.) He stressed that he did not want a mixture of computer scietists and biologists, he wanted scientists with a mixture of computing and biology. Since his future involves molecules, maybe he’s also hiring chemist/computerScientists…
But we are actively discouraging the sort of work envisioned By Lederberg and Corey 30 years ago. There are exceptions – I spent 3 hours with my colleague Steve Ley discussing how we can bring modern informatics into synthetic chemistry. I am sure that our biggest problem is the lack of an immediate Open global knowledge base in chemistry. It’s all there on paper, but to get it into a machine is a mighty task. It will need new methods of computing – including social computing and I’ll explore these ideas systematically in this blog. We might even achieve something with your help.
So I am pleased to see the quality of the chemical blogs, even if Tenderbutton is retiring. With lightweight mashup-like approaches we may be able to use the new approaches to informatics that are being developed in social computing. Biology has control of its knowledgebase – it had to fight to keep it in the genome information wars- but it’s vibrant and innovative. Chemistry has surrendered its knowledgebase to commercial and quasi-commercial interests who point in the direction of pharma rather than the information revolution. I will show in a week or two how we might be able to start regaining some of it.
P.
-
Recent Posts
-
Recent Comments
- pm286 on ContentMine at IFLA2017: The future of Libraries and Scholarly Communications
- Hiperterminal on ContentMine at IFLA2017: The future of Libraries and Scholarly Communications
- Next steps for Text & Data Mining | Unlocking Research on Text and Data Mining: Overview
- Publishers prioritize “self-plagiarism” detection over allowing new discoveries | Alex Holcombe's blog on Text and Data Mining: Overview
- Kytriya on Let’s get rid of CC-NC and CC-ND NOW! It really matters
-
Archives
- June 2018
- April 2018
- September 2017
- August 2017
- July 2017
- November 2016
- July 2016
- May 2016
- April 2016
- December 2015
- November 2015
- September 2015
- May 2015
- April 2015
- January 2015
- December 2014
- November 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- February 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- August 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- May 2007
- April 2007
- December 2006
- November 2006
- October 2006
- September 2006
-
Categories
- "virtual communities"
- ahm2007
- berlin5
- blueobelisk
- chemistry
- crystaleye
- cyberscience
- data
- etd2007
- fun
- general
- idcc3
- jisc-theorem
- mkm2007
- nmr
- open issues
- open notebook science
- oscar
- programming for scientists
- publishing
- puzzles
- repositories
- scifoo
- semanticWeb
- theses
- Uncategorized
- www2007
- XML
- xtech2007
-
Meta
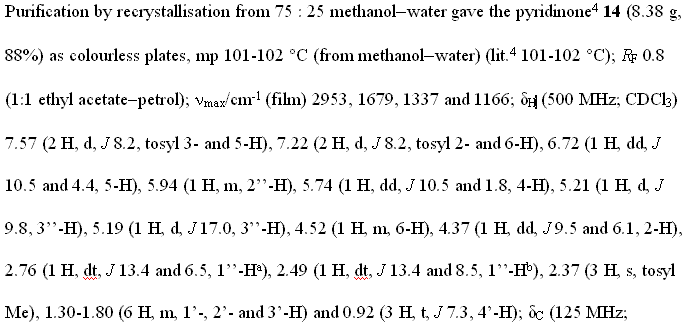

 Alert level C
Alert level C