Chemspider has recently replied in depth to my concerns:
ChemSpider as a part of Web 2.0 – and what is that Web 2.0 anyways?
In this blog I am going to excerpt from another blog (and bolded to identify) regarding ChemSpider (based on my previous post it’s the way of the blogosphere) and it’s non Web 2.0 status since pages from the ChemSpider blog are being excerpted in the same way.
I shall tackle the Web 2.0 issues separately but here I am concerned that the material and services produced by Chemspider are likely to be seriously misused by students. Chemspider states:
May I use your service in my teaching class ?
Absolutely. We would especially like the academic community to benefit from the information available on ChemSpider.
Now students – especially those startiing their courses – are likely to accept what they read on the web. When they access a company who sells the calculation of molecular properties the assume that the molecules, calculation and metadata is of sufficient quality for them to use. They cannot be expected to have enough judgment to assume that a large number of the answers they get will be wrong, and that the definitions and explanations of the properties are wrong or unclear. While I, as an experienced scientists in chemoinformatics realise how suspect much of the material and services (not just from Chemspider) is suspect, students cannot.
Here are the “definitions” of some of the properties that Chemspider/ACD provide. The standard of description, the lack of units and metadata, and experimental constraints would be below that that an undergraduate would be expected to present: I shall pick out a few examples
PhysChem Properties (as defined by ACD/Labs):
The Partition Coefficient (LogP) is the equilibrium distribution of a solute between two liquid phases, the constant ratio of the solute’s concentration in the upper phase to its concentration in the lower phase. ACD/Labs provides acess to logP prediction through their freeware ACD/ChemSketch and their LogP addon.
PMR: The Partition Coefficient is NOT LogP, it is P.
The Distribution Coefficient (LogD) is the ratio of the amounts of solute dissolved in two immiscible liquids at equilibrium. The distribution coefficient (logD) equation accounts for all possible partition coefficients (logP) that a system can obtain. For compounds containing a single ionizable group (acid/base) there are 2 partition coefficients or a single distribution coefficient accounting for the relative concentration of each species within each of the two possible phases
PMR: these two paragraphs do not make it clear what the differences between D and P (or LogD and LogP). Moreover there is no mention that a P (or LogP) is meaningless unless the solvents are fully identified. (I assume that the non-polar phase is octanol but I cannot find this in this document or in the calculation of properties. As P is temperature dependent it is necessary to report this – but I cannot find this in the data. (I assume it is 298K but this is not mentioned)
Polar Surface Area (PSA) is the measure of how much exposed polar area any two- or three-dimensional object has.
PMR: This is a fuzzy definition – the algorithm calculates a precise quantity but this gives no indication of how this is done. Different vendors will report different values for this quantity. The impression given is that the precise definition is unimportant. Note also that when this property is first displayed to the student there are no units (we try very hard to impress on students that all numeric quantities must have units).
Surface Tension is a property of liquids arising from unbalanced molecular cohesive forces at or near the surface, as a result of which the surface tends to contract and has properties resembling those of a stretched elastic membrane
PMR: .Again a fuzzy defintion that many educators would indicate that the student did not understand surface tension.
Molar Refraction is the equation for the refractive index of a compound modified by the compound’s molecular weight and density. Also known as the Lorentz-Lorenz molar refraction.
PMR: Molar Refraction is NOT an equation. Any student writing stuff like this would get near-zero marks.
Now I appreciate that Chemspider is “beta” which means that they want to community to correct their bugs but it is not fair to encourage students to be part of it. For example if you want properties of “sodium hydride” it will draw a picture looking like:
Na+ HH2–
It is clear that this is NOT a copy of the pubchem entry (which is the normal NaH) but that the Chemspider software (or the ACD software) has taken a correct formula and displayed the formula incorrectly. Verify this for yourself but do not let students near it
Chemspider will calculate properties for any compound, and many of these are meaningless. For example, try “prussian blue” and it will give a logP even though the stuff is an insoluble pigment. Now that is because the chemical formula has been represented as separated iron ions and cyanide ions. This may be useful for searching, but it unacceptable for calculating properties.
So, in summary, you cannot rely on some of the properties calculated by Chemspider. For students that means you should not rely on any.
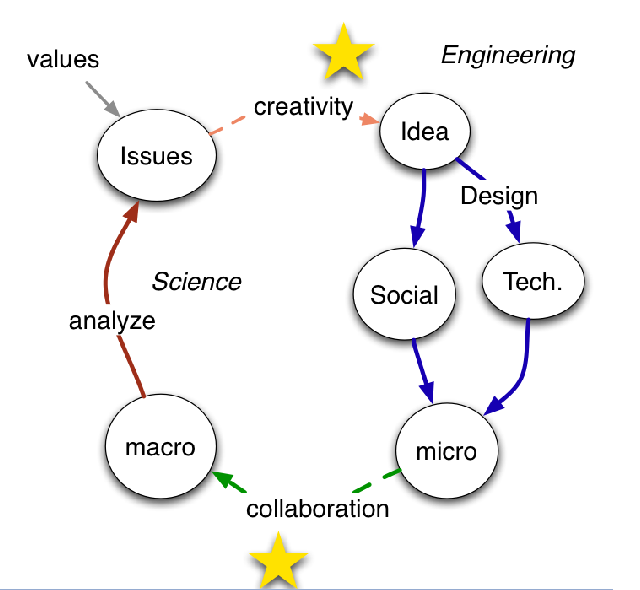
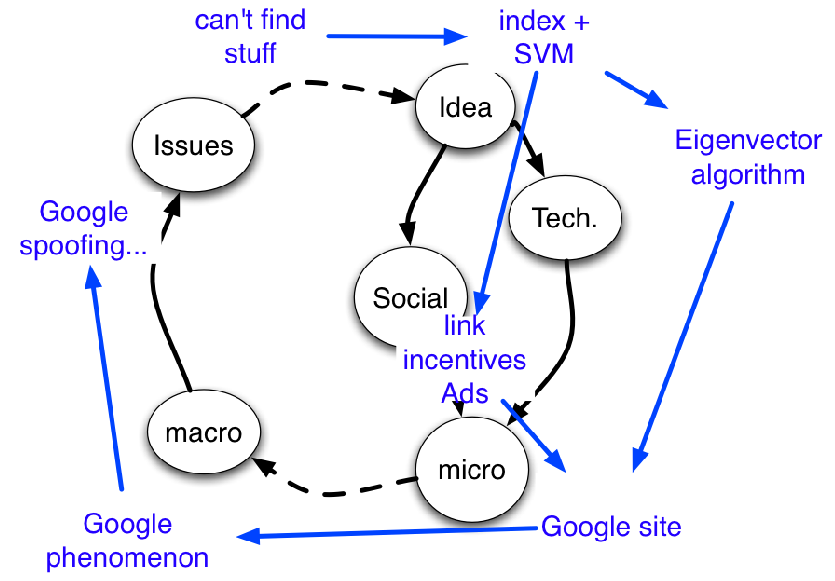
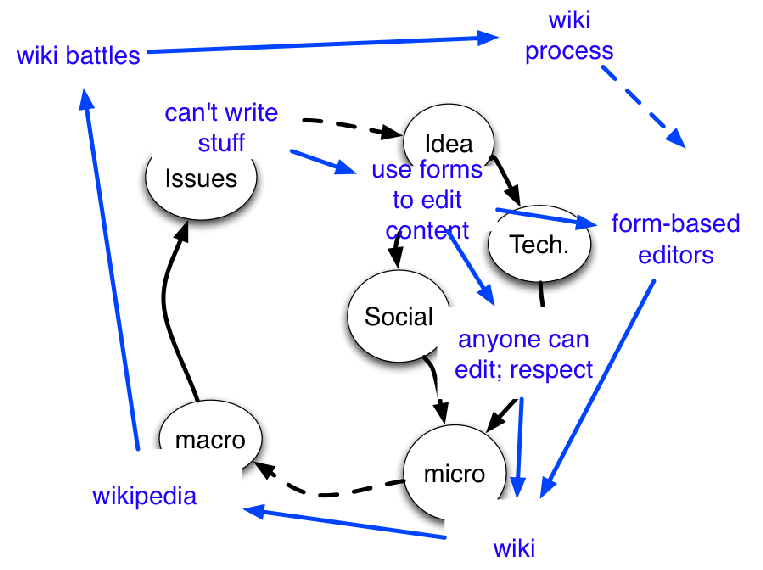
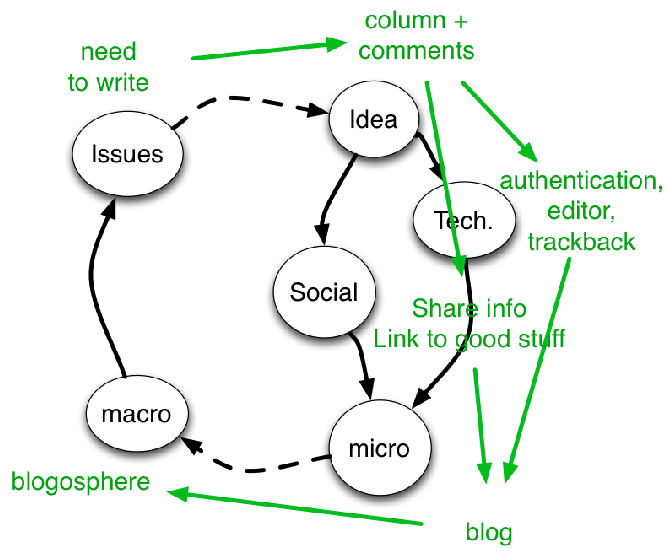

 But I want to know more… maybe the data need re-interpreting … so let’s read the whole article …
But I want to know more… maybe the data need re-interpreting … so let’s read the whole article …

May 11th, 2007 at 7:45 pm eAs far as I can tell, there are around 3000 compounds with chemboxes, and over 2000 with drugboxes. I think we have many compounds on WP without chemboxes, but they are typically very brief articles (stubs) with little information. Of course linking into the mainstream of chemical information, as dbpedia seeks to do, may provide an incentive for more wikichemists to work on adding chemboxes. Sounds great!
Martin A. Walker (Walkerma on WP)