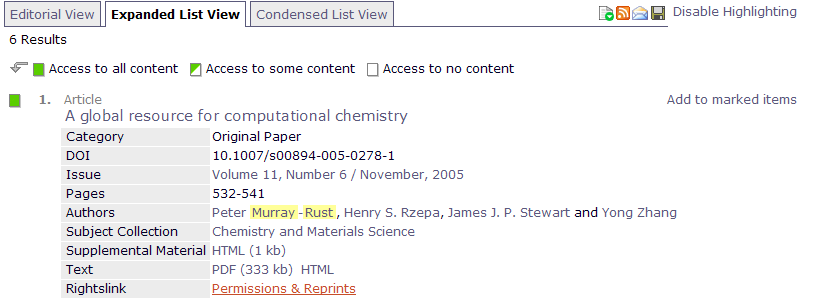
Yesterday I needed the measured (i.e. not predicted) mass density for 2-bromo-propanoyl-bromide (CH3-CH(Br)C(=O)Br). This is a moderately common reagent and so I went to look for it on the Web – ultimately finding it on several sites. The value is ca. 2.061 g.cm-3 (many sites omit the units – argh!!). The temperature should also be reported – but isn’t. I need the measured density because many chemical recipes give the volume of reagents and I want to work out the molar ratios in reactions for which I need the density. I may also be interested in other measured properties such as boiling point.
The problem is that it’s difficult to scrape these sites. They give little indication of copyright, are arcanely structured and often have poor semantics (e.g. units). The best known is the NIST Webbook, part of which reads:
-
Thermophysical property data for 74 fluids:
- Density, specific volume
- Heat capacity at constant pressure (Cp)
- Heat capacity at constant volume (Cv)
- Enthalpy
- Internal energy
- Entropy
- Viscosity
- Thermal conductivity
- Joule-Thomson coefficient
- Surface tension (saturation curve only)
- Sound speed
You can search for data on specific compounds in the Chemistry WebBook based on name, chemical formula, CAS registry number, molecular weight, chemical structure, or selected ion energetics and spectral properties.
NIST reserves the right to charge for access to this database in the future.The National Institute of Standards and Technology (NIST) uses its best efforts to deliver a high quality copy of the Database and to verify that the data contained therein have been selected on the basis of sound scientific judgment. However, NIST makes no warranties to that effect, and NIST shall not be liable for any damage that may result from errors or omissions in the Database.
© 1991, 1994, 1996, 1997, 1998, 1999, 2000, 2001, 2003, 2005 copyright by the U.S. Secretary of Commerce on behalf of the United States of America. All rights reserved.
It’s clear that this is not an Open site – most works of the US Government are required to make their works freely available but NIST has exemption for its databases so that it can raise money.
Many suppliers list property information but scattered throughout somewhat uncoordinated pages. Moreover the copyright and crawling position is often not clear.
My requirement is likely to be via robot – i.e. an asynchronous request for a property I don’t have, with the ability to re-use it without explicit permission. I am therefore wondering whether there are Open sites for chemical data that can be accessed without explicit permission. I am not interested in collections of millions of compounds, but rather ca. 10,000 of the most commonly used.
A good source of data is MSDS (Materials Safety data Sheets), and here is part of a typical one hosted by a group at Oxford University:
General
Synonyms: nitrilo-2,2′,2″-triethanol, tris(2-hydroxyethyl)amine, 2,2′,2″-trihydroxy-triethylamine, trolamine, TEA, tri(hydroxyethyl)amine, 2,2′,2″-nitrilotrisethanol, alkanolamine 244, daltogen, sterolamide, various further trade names
Molecular formula: C6H15NO3
CAS No: 102-71-6
EC No: 203-049-8
Physical data
Appearance: viscous colourless or light yellow liquid or white solid
Melting point: 18 – 21 C
Boiling point: 190 – 193 C at 5 mm Hg, ca. 335 C at 760 mm Hg (decomposes)
Vapour density: 0.01 mm Hg at 20 C
Vapour pressure: 5.14
Specific gravity: 1.124
Flash point: 185 C
Explosion limits: 1.3 % – 8.5 %
Autoignition temperature: 315 C
Stability
Stable. Incompatible with oxidizing agents and acids. Light and air sensitive.
It looks as if there are in the range of 5,000 to 100,000 compounds on the site – I haven’t counted and if so this is close to what I am looking for. It looks as if the creators are happy for people to download it – their concern is that it shouldn’t be seen as authoritative about safety (a perfectly reasonable request). If so, an Open Data sticker would be extremely useful and solve the problem. (There is the minor problem that there are no connection tables, but links to Pubchem should solve that).
There has been talk of a Wikichemicals – and this is the sort of form it might take. It shouldn’t be too difficult to create it and the factual data on the pages doesn’t belong to anyone. So I’d like to know whether anyone has been doing this (measured, not predicted data) and whether there resource is Open.