Nature has just published an account of Chemspider after interviewing a number of people. (The Nature report, Geoff Brumfiel and I spent considerable time on the phone but it was too late to include my comments in the article so I” outline what I think I may have said at the bottom – I have tried to be objective and am reviewing CS as it is now. Many of my comments relate to chemical aggregators in general. I include the whole of Antony’s post as, presumably, I will get into trouble if I copy very much from Nature.
22:32 07/05/2008, Antony Williams,
Last a week I had a pleasant chat with a reporter from Nature magazine, a Mr Geoff Brumfiel. Geoff was interested in ChemSpider…what it was, how it ran, who used it, who supported it, who liked it, who curated it, who didn’t like it and so on.
The results of that discussion, and others he spoke to about ChemSpider, are here in his article.
Chemists spin a web of data p139
Chemspider website provides free information on millions of molecules.
Geoff Brumfiel
doi:10.1038/453139a
Full Text | PDF
It is a rule at Nature, at least for this type of article, that I [AJW] could not see the article before it went to press and therefore I didn’t get the chance to proofread and comment. Geoff has accurately captured the spirit of our discussions but a few detailed clarifications are needed too. I have pasted in black the article content and in italics the clarification.
“
providing the community with an open-access source of chemical information”
I giggled and commented please don’t say it’s Open Access. Say it’s Free Access. Say there are Open Data. And now we have Creative Commons licenses. But don’t say it’s Open Access, not Strong, not weak, not gold, not green. Just Free Access. No price barriers to usage.
“
Chemist Antony Williams is hoping to change this in a move likely to ruffle the feathers of the American Chemical Society.”
I commented that we are not purposely in competition with anyone. It’s not what drives us to do this. Whether others see us to be competitive is for them not us. We don’t intentionally try to ruffle feathers. It doesn’t mean that what we are doing won’t ruffle feathers of course. Whether it’s ACS or others. It’s not the goal..it might be an outcome.
“
The modest project has made chemists interested in open access take notice — last week, the number of daily users of the site surpassed 5,000.”
We have crossed 5500 users for the past two nights. The trend is positive.
“Other potential sources of information, such as Wikipedia, lack the algorithms needed to search chemicals according to their structure. “
Structure searching is “feasible” of course with InChI Strings. But substructure isn’t and Wikipedia is treated as a text-based search by almost all of its users
“The site is maintained with modest profits from advertising and the work of about 30 active volunteers who double-check the data pulled in from outside.”
The original investment in hardware and software costs has finally been recouped. Modest profits? No one gets paid for the work we do. There is a phenomenal sweat equity investment in the platform numbering many thousands of hours to get here. We are indebted to the many software collaborators, providers of tools and the people curating and depositing to the system. There have BEEN about 30 active volunteers. RIght now I would say the number of active depositors and curators is around 10. But it is growing. I hadn’t checked the number of REGISTERED users for a long time. We have over 1150 registered users…those who CAN login and curate data, deposit data, see new features etc. People do NOT have to register to use the site…but >1150 did. Wow. I didn’t know it was that many until i just checked (BIG SMILE)
““There’s an awful lot of chemical information, but there’s an awful lot of rubbish as well,” says Barrie Walker, a retired industrial chemist in Yorkshire, UK, who helps maintain the site.”
Don’t know whether Marrie said this or not. He IS an honest guy and he is our QUALITY GURU and we are proud that he is willing to give us his fine eyes. There IS garbage on the site still. But, after a year online and active curating it has been much reduced. About 200 edits a day are made to the site: names changed/deleted/added, spectra/structures/URLs/Publications added etc. It’s quite the pace. We have cleaned up 100s of thousands of incorrect associations from the external data sources. It’s been and will remain an enormous task with an enormous payback for the community
“
Williams adds that the site still has problems with certain searches. For example, it struggles to distinguish between isomers: molecules with the same chemical formula arranged in different structures.”
We can distinguish isomers no problem. The PROBLEM is that there is a mixture of isomeric species submitted from multiple data sources and data are mixed and intermingled in way that the user cannot get to the correct structure. Search taxol or Ginkgolide on the ChemSpider blog and read the mutliple blog posts about this. We can of course search all isomers for a particular chemical formula…
“But Williams nevertheless believes that the service may be able to compete with for-profit services. “What I’m doing is highly disruptive,” he says. “I think it can be done and it needs to be done.””
I think what WE are doing…its not me..it’s we…is disruptive. In a good way. Many chemists will benefit. Will it have an impact on for-profit services? Yes, maybe. As an outcome but not as the target. Our team of people, both internal to ChemSpider’s development and Advisory Group, and the people we don’t even know who are cleaning and depositing into the system for their colleagues in the community, are creating a powerful resource for Chemists. The FOCUS of this effort is to Build a Structure Centric Community for Chemists. We will change that soon…the focus on Structure-Centric will be to cover Chemistry in general and to Build a Community for Chemists.
We are well on our way and thanks to Nature, and Geoff in particular for exposing it. My comments above are not meant to detract from Geoff’s reporting abilities but it was a long discussion and some clarification statements are of value i believe.
PMR; Firstly to say that I commented to Geoff before Chemspider’s announcement that it was adopting CC-SA licences. This is a major advance and has enhanced the importance of Chemspider. For non-chemists the lack of data in chemistry is a desperate and desperately serious problem. Almost all publicly visible data is first published in peer-review journals. (There are exceptions, where data is collected for hire, and I have no problem with people charging for that – it is the charging for data that belongs to the community that concerns me. So, in challenging the status quo, Chemspider is pointing in the right direction).
It’s (now) based on Web 2.0 principles in that it uses social computing for some of its content and can and has reacted to external changes. It’s also perpetual beta. It’s not, however, based on semantic web technology such as RDF and XML and this may be a future limitation in managing some of the more complex content. Although I’m not party to the internal design I’d guess it has a relational database, most of whose primary keys are the identifiers for chemical compounds. These identifiers map onto canonicalised chemical structures (one serialization of which is the InChI) and this is the primary mechanism for indexing compounds. Chemistry is fortunate in that it is easier to index compounds automatically than, say, stellar objects, organisms, genomes, etc.
The information management is hybrid. At one level there is robotic ingestion and curation and at the other human annotation (curation). CS has ca 20 million compounds and the only way to manage these is robotically. This brings several problems, which bedevil any large chemical aggregator:
- the data either have to come from somewhere else or be computer-generated. CS does both – it ingests from PubChem, and it computes molecular properties. Pubchem (which I’ll tackle in a later post) consists mainly of data contributed by a number of parties and is of highly variable quality. It is extremely difficult to evaluate this sort of data robotically as there are few objective constraints and few other independent data sources. (We are trying to do this for a much smaller data set ~ 5000 compounds, and we find unexpected and serious garbage, which I’ll blog later).
- Similarly there is no guarantee that the computation of properties is free from error – indeed it cannot be. Many physical properties depend on the physical form of the compound and this is often not recorded. I suspect most of the properties are computed by heuristic means (“QSPR”) rather than QM calculations. And many of them fail to take things like chemical stability and reactivity into account. (Examples are boiling points for compounds that decompose, flashpoints for things that could never burn). But how do you tell this robotically – I don’t have a good suggestion But one can guarantee that in 20 million calculations some will be meaningless
- Chemistry is not regular, and in millions of compounds there will be mainly that simply don’t behave as expected from their formula. Or, alternatively, some can have many formulae. There is no simple robotic way of determining which these are and correcting them. So the compiler and the user of such systems have to be clear that error is part of the nature of the system.
Chemspider is using social computing (crowdsourcing) to clean up (curate) the information in the database. This works in Wikipedia, although the number of chemicals in in the thousands, not the millins, and there are still many data and chemical problems. Moreover WP shows that there are compounds – e.g. aluminium chloride – where there is no single structure. It’s a matter of opinion as to whether the various states are manifestations of a single compound or several separate ones – certainly they have different connections tables. The problem with crowdsourcing is the numbers – chemistry is conservative – chemical WP lags behind other science, despite enormous efforts from a small number of individuals including Antony. What is certain is that crowdsourcing can only address a very small amount of Chemspider content – even with 10000 volunteers it would take 2000 curations each to address the whole.
Chemspider has also started to act as a repository for scientific data, especially Jean-Claude Bradley’s Open Notebook Science. In doing that it runs up against the same problems as University Institutional Repositories – heterogeneous data sets, versioning, metadata, compound documents, etc. Its advantage is that it will probably be restricted to a fairly narrow range of content types (chemistry) and it is also able to provide chemical substructure search (a major problem across the web
What is Chemspider now is and where it may be going? It’s difficult to predict anything on the web but it’s also clear that chemists are one of the most conservative disciplines. Why use a free service when you can get your library to pay (a lot of money) for ACS or Beilstein services? So I wouldn’t predict explosive growth like Flickr or Google.
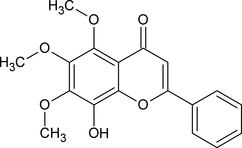
I think quality is a major problem in this area. Chemspider is correcting the structures of compounds as they come across errors. In some cases this is possible as “all chemists know the correct structure of X”. But in many cases they won’t agree, or the chemistry simply doesn’t admit of an answer. There is no correct structure for glucose. And then there is the problem of the long tail. There’s a huge amount of chemistry out there and a lot is wrong. When InChI started, Nick Day surveyed the web for chemical structures that InChI might help to disambiguate. We started with staurosporine – it’s an anticancer drug that one of my close associates was interested in and it wasn’t clear what the structure was. Nick found 26 sites displaying staurosporine and there were 19 different structures given. Some were incomplete and several were just crazily wrong. Clearly many chemical suppliers, journal editors, etc. do not care about chemical structures. So there is a huge amount of rubbish out there.
However as Nature says:
Chemical data have long been available, but at a hefty price. The largest supplier of such information is the American Chemical Society’s Chemical Abstracts Service. The service, which is more than a century old, includes data on roughly 35 million molecules. But university and industry chemists must pay thousands of dollars to use the database. The society will not reveal numbers, but fees for using the database are thought to make up a substantial portion of its US$311-million annual income from ‘electronic services’. Some have been highly critical of the society’s grip on chemicals.
PMR: At some stage, therefore, the community will react against this centralisation of information, but it could be a long time. I don’t think anyone should set up to duplicate what ACS does – I think we should use modern thinking to do things quicker, smarter, cheaper and in tune with the modern Web. Chemspider may have to make some choices soon – is it a company or a voluntary activity? does it concentrate on high volume and variable quality, or low volume and high quality – it cannot do both? What is the particular USP of its repository service ?- there may well be a role for a specialist chemical repository service but when? Is it different from Pubchem, and how…?