Typed into Arcturus
The first pass of the automatic extraction of chemical information from patents is going well on a mechanical level.
- One weekly index has 30-200 appropriate patents. Each has between 0 and 1500 images of chemical relevance
- Each index therefore has ca 10,000 images, almost all of chemical compounds or general formulae or reactions.
- We use OSRA (Open Source, NIH) to interpret the images. It takes about 1-30 secs each and the first index will complete in ca 24 hours. This means that we could do this task for the last 10 years in 500 distributed days. I’d like to do that before #solo10. (I could do it all at Cambridge, but I’d rather it were citizen-science.)
So far the “record” is a patent with 1500 images. Here’s one (EP_2050749A1/0026imgb0032.tif)
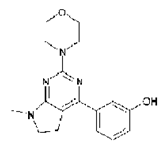
Could someone please tell me what the InChI or SMILES or CML is for this compound?
I am now working on the text-mining. More later today.
Not found in ChemSpider… add it to ChemPedia though:
http://chempedia.com/substances/0-4501-4228-3902
Will draw it now in Bioclipse to get SMILES, InChI, and CML.
Mmm… addED … BTW, did they give it a name? If so, please add it to the ChemPedia entry… you can create an account with any OpenID…
InChI=1S/C17H22N4O2/c1-20-8-7-14-15(12-5-4-6-13(22)11-12)18-17(19-16(14)20)21(2)9-10-23-3/h4-6,11,22H,7-10H2,1-3H3
InChIKey: UNGDZCFSJUIHHY-UHFFFAOYSA-N
SMILES: OC=1C=CC=C(C=1)C=2N=C(N=C3C=2CCN3C)N(C)CCOC
CML:
C.sp2
N.sp2
C.sp2
C.sp2
C.sp2
N.sp2
C.sp2
N.sp3
C.sp3
C.sp3
C.sp3
O.sp3
C.sp3
N.sp3
C.sp3
C.sp3
C.sp3
C.sp2
C.sp2
C.sp2
C.sp2
C.sp2
O.sp3
OC=1C=CC=C(C=1)C=2N=C(N=C3C=2CCN3C)N(C)CCOC
OK, now escaped:
<?xml version=”1.0″ encoding=”ISO-8859-1″?>
<list convention=”cdk:model” id=”model1″ xmlns=”http://www.xml-cml.org/schema”>
<moleculeList convention=”cdk:moleculeSet” id=”molSet1″>
<molecule id=”m1″>
<atomArray>
<atom id=”a1″ elementType=”C” x2=”-2.082499999999998″ y2=”2.0125″ formalCharge=”0″ hydrogenCount=”0″>
<atomType convention=”bioclipse:atomType”>C.sp2</atomType>
</atom>
<atom id=”a2″ elementType=”N” x2=”-0.8700644347017847″ y2=”1.3124999999999982″ formalCharge=”0″ hydrogenCount=”0″>
<atomType convention=”bioclipse:atomType”>N.sp2</atomType>
</atom>
<atom id=”a3″ elementType=”C” x2=”-0.8700644347017867″ y2=”-0.0875000000000018″ formalCharge=”0″ hydrogenCount=”0″>
<atomType convention=”bioclipse:atomType”>C.sp2</atomType>
</atom>
<atom id=”a4″ elementType=”C” x2=”-2.0825000000000014″ y2=”-0.7875000000000002″ formalCharge=”0″ hydrogenCount=”0″>
<atomType convention=”bioclipse:atomType”>C.sp2</atomType>
</atom>
<atom id=”a5″ elementType=”C” x2=”-3.294935565298215″ y2=”-0.08749999999999925″ formalCharge=”0″ hydrogenCount=”0″>
<atomType convention=”bioclipse:atomType”>C.sp2</atomType>
</atom>
<atom id=”a6″ elementType=”N” x2=”-3.294935565298214″ y2=”1.3125000000000004″ formalCharge=”0″ hydrogenCount=”0″>
<atomType convention=”bioclipse:atomType”>N.sp2</atomType>
</atom>
<atom id=”a7″ elementType=”C” x2=”0.34237113059642654″ y2=”-0.7875000000000035″ formalCharge=”0″ hydrogenCount=”0″>
<atomType convention=”bioclipse:atomType”>C.sp2</atomType>
</atom>
<atom id=”a8″ elementType=”N” x2=”-2.0824999999999965″ y2=”3.4125000000000005″ formalCharge=”0″ hydrogenCount=”0″>
<atomType convention=”bioclipse:atomType”>N.sp3</atomType>
</atom>
<atom id=”a9″ elementType=”C” x2=”-3.2949355652982097″ y2=”4.1125000000000025″ formalCharge=”0″ hydrogenCount=”3″>
<atomType convention=”bioclipse:atomType”>C.sp3</atomType>
</atom>
<atom id=”a10″ elementType=”C” x2=”-0.8700644347017819″ y2=”4.1125″ formalCharge=”0″ hydrogenCount=”2″>
<atomType convention=”bioclipse:atomType”>C.sp3</atomType>
</atom>
<atom id=”a11″ elementType=”C” x2=”-0.8700644347017817″ y2=”5.5125″ formalCharge=”0″ hydrogenCount=”2″>
<atomType convention=”bioclipse:atomType”>C.sp3</atomType>
</atom>
<atom id=”a12″ elementType=”O” x2=”-2.082499999999996″ y2=”6.2125″ formalCharge=”0″ hydrogenCount=”0″>
<atomType convention=”bioclipse:atomType”>O.sp3</atomType>
</atom>
<atom id=”a13″ elementType=”C” x2=”-3.2949355652982106″ y2=”5.5125″ formalCharge=”0″ hydrogenCount=”3″>
<atomType convention=”bioclipse:atomType”>C.sp3</atomType>
</atom>
<atom id=”a14″ elementType=”N” x2=”-4.335338320966568″ y2=”-1.0242828489024″ formalCharge=”0″ hydrogenCount=”0″>
<atomType convention=”bioclipse:atomType”>N.sp3</atomType>
</atom>
<atom id=”a15″ elementType=”C” x2=”-3.7659070206604484″ y2=”-2.3032464896020417″ formalCharge=”0″ hydrogenCount=”2″>
<atomType convention=”bioclipse:atomType”>C.sp3</atomType>
</atom>
<atom id=”a16″ elementType=”C” x2=”-2.3735763671448655″ y2=”-2.156906641027328″ formalCharge=”0″ hydrogenCount=”2″>
<atomType convention=”bioclipse:atomType”>C.sp3</atomType>
</atom>
<atom id=”a17″ elementType=”C” x2=”-5.704744961993896″ y2=”-0.7332064817575359″ formalCharge=”0″ hydrogenCount=”3″>
<atomType convention=”bioclipse:atomType”>C.sp3</atomType>
</atom>
<atom id=”a18″ elementType=”C” x2=”1.5548066958946425″ y2=”-0.08750000000000402″ formalCharge=”0″ hydrogenCount=”1″>
<atomType convention=”bioclipse:atomType”>C.sp2</atomType>
</atom>
<atom id=”a19″ elementType=”C” x2=”2.7672422611928553″ y2=”-0.7875000000000058″ formalCharge=”0″ hydrogenCount=”0″>
<atomType convention=”bioclipse:atomType”>C.sp2</atomType>
</atom>
<atom id=”a20″ elementType=”C” x2=”2.7672422611928535″ y2=”-2.1875000000000058″ formalCharge=”0″ hydrogenCount=”1″>
<atomType convention=”bioclipse:atomType”>C.sp2</atomType>
</atom>
<atom id=”a21″ elementType=”C” x2=”1.5548066958946387″ y2=”-2.887500000000004″ formalCharge=”0″ hydrogenCount=”1″>
<atomType convention=”bioclipse:atomType”>C.sp2</atomType>
</atom>
<atom id=”a22″ elementType=”C” x2=”0.34237113059642565″ y2=”-2.187500000000003″ formalCharge=”0″ hydrogenCount=”1″>
<atomType convention=”bioclipse:atomType”>C.sp2</atomType>
</atom>
<atom id=”a23″ elementType=”O” x2=”3.979677826491069″ y2=”-0.0875000000000068″ formalCharge=”0″ hydrogenCount=”1″>
<atomType convention=”bioclipse:atomType”>O.sp3</atomType>
</atom>
</atomArray>
<bondArray>
<bond id=”b1″ atomRefs2=”a1 a2″ order=”D”>
<bondType dictRef=”cdk:aromaticBond”/>
</bond>
<bond id=”b2″ atomRefs2=”a2 a3″ order=”S”>
<bondType dictRef=”cdk:aromaticBond”/>
</bond>
<bond id=”b3″ atomRefs2=”a3 a4″ order=”D”>
<bondType dictRef=”cdk:aromaticBond”/>
</bond>
<bond id=”b4″ atomRefs2=”a4 a5″ order=”S”>
<bondType dictRef=”cdk:aromaticBond”/>
</bond>
<bond id=”b5″ atomRefs2=”a5 a6″ order=”D”>
<bondType dictRef=”cdk:aromaticBond”/>
</bond>
<bond id=”b6″ atomRefs2=”a6 a1″ order=”S”>
<bondType dictRef=”cdk:aromaticBond”/>
</bond>
<bond id=”b7″ atomRefs2=”a3 a7″ order=”S”/>
<bond id=”b8″ atomRefs2=”a1 a8″ order=”S”/>
<bond id=”b9″ atomRefs2=”a8 a9″ order=”S”/>
<bond id=”b10″ atomRefs2=”a8 a10″ order=”S”/>
<bond id=”b11″ atomRefs2=”a10 a11″ order=”S”/>
<bond id=”b12″ atomRefs2=”a11 a12″ order=”S”/>
<bond id=”b13″ atomRefs2=”a12 a13″ order=”S”/>
<bond id=”b14″ atomRefs2=”a5 a14″ order=”S”/>
<bond id=”b15″ atomRefs2=”a14 a15″ order=”S”/>
<bond id=”b16″ atomRefs2=”a15 a16″ order=”S”/>
<bond id=”b17″ atomRefs2=”a16 a4″ order=”S”/>
<bond id=”b18″ atomRefs2=”a14 a17″ order=”S”/>
<bond id=”b19″ atomRefs2=”a18 a19″ order=”D”>
<bondType dictRef=”cdk:aromaticBond”/>
</bond>
<bond id=”b20″ atomRefs2=”a19 a20″ order=”S”>
<bondType dictRef=”cdk:aromaticBond”/>
</bond>
<bond id=”b21″ atomRefs2=”a20 a21″ order=”D”>
<bondType dictRef=”cdk:aromaticBond”/>
</bond>
<bond id=”b22″ atomRefs2=”a21 a22″ order=”S”>
<bondType dictRef=”cdk:aromaticBond”/>
</bond>
<bond id=”b23″ atomRefs2=”a22 a7″ order=”D”>
<bondType dictRef=”cdk:aromaticBond”/>
</bond>
<bond id=”b24″ atomRefs2=”a7 a18″ order=”S”>
<bondType dictRef=”cdk:aromaticBond”/>
</bond>
<bond id=”b25″ atomRefs2=”a19 a23″ order=”S”/>
</bondArray>
<scalar dictRef=”cdk:molecularProperty” title=”net.bioclipse.cdk.domain.property.SMILES” dataType=”xsd:string”>OC=1C=CC=C(C=1)C=2N=C(N=C3C=2CCN3C)N(C)CCOC</scalar>
</molecule>
</moleculeList>
</list>
Peter, I guess this is the compound:
http://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=24888923
When seeing the image for the first time, I had the feeling a bond was too thin to show up in the rasterized image… this hit on PubChem might be a lead for further information.