Rich Apodaca has asked me to show how CML can deal with metallocene compounds – and I’m happy to do this – it comes at a very good time. He points to Metallome blog and I’ll copy some of the material on ferrocene. I’ll show the post and then explain the approach
Metallome: Drawing ferrocene
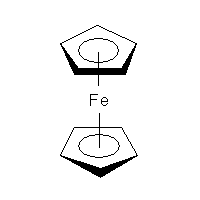
Ferrocene was discovered in 1951 and we still do not know the proper way to draw it. CrossFire example recommends to connect every carbon atom of the ring to the central metal atom. Which is fair enough and will be a valid query for CrossFire Gmelin database. Similarly, both ChEBI and NIST Webbook use decacoordinate iron in ferrocene structure (a). In this representation, all carbon—carbon bonds are single. But, according to IUPAC Recommendations, section GR-1.7.2,coordination bonds to contiguous atoms (most commonly representing a form of π-bonding) should be drawn to indicate most clearly that special bonding pattern. Depictions that imply a regular covalent bond — and especially, depictions that show a regular covalent bond to each member of a delocalized system — are not acceptable.
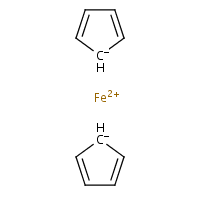
In other words, the preferred representation is the one with bicoordinate iron and delocalised bond system (b). The problem with that is there is no agreed (as far as chemoinformaticans are concerned) way to do that, even though solutions for different applications (e.g. for Marvin Sketch) do exist. In MolBase, the coordination number of iron in ferrocene is 6 (and I do remember Mark Winter confirming that this is true). On yet another hand, Beilstein and ChemIDplus databases represent ferrocene as a standalone Fe2+ and two standalone cyclopenta-2,4-dienide anions (c), thus avoiding the question of coordination number altogether. Naturally, the decacoordinate-iron query will not work in Beilstein. (For InChI implications, see this discussion.)
(a)
(b)
(c)
PMR: Many thanks Kirill for this very clear explanation. The first and central point is that there is no agreed way to represent ferrocene, and the semantic approach honours this.
in CML we represent what we do know, and do that as fully as possible. Implicit semantics (i.e. information that has to be provided by the reader or reading program) creates enormous problems. A typical example of implicit semantics is omitting hydrogen atoms and although CML allows this we are not allowing omitted H in Chem4Word.
So let’s build up systematically. What do we know? We know we have a molecule (ferrocene exists in the gas phase so we can talk of single molecules and don’t have to worry about substances at this stage).
<molecule id="mol123456789" title="ferrocene"
xmlns='http://www.xml-cml.org/schema'/>
What an anticlimax! we knew that. But we have at least told the world we have a molecule. Let’s see what the world has to offer… off to Pubchem for Ferrocene search. This gives a huge amount of enties and shows the chaos when we don’t have semantic chemistry – several entries have a formula of MF: C10H10Fe-6 which is obviously caused by a non-semantic program trying to work out the formula from non-semantic input.
The lesson is simple: If you care about quality and validity you must use a semantic approach.
So how do we know which is actually “ferrocene”. Simple answer – we don’t. We have a number of conflicting pieces of information – some have a name “ferrocene” but the formula associated with those names varies. Of course we as inorganic chemists know the “correct” formula, but it still means Pubchem (and much else) doesn’t act as a simple lookup. We have to add metadata – who asserted what. That’s where semantics starts to come in.
We’ll take the first entry:
| SID: 49854569 | <!– var Menu49854569_1 = [ [“UseLocalConfig”, “jsmenu3Config”, “”, “”], [“Same Substances” , “window.top.location=’/sites/entrez?Db=pcsubstance&DbFrom=pcsubstance&Cmd=Link&LinkName=pcsubstance_pcsubstance_same_popup&LinkReadableName=Same%20Substances&IdsFromResult=49854569&ordinalpos=1&itool=EntrezSystem2.PEntrez.Pcsubstance.Pcsubstance_ResultsPanel.Pcsubstance_RVDocSum’ “, “”, “”], [“Same Parent” , “window.top.location=’/sites/entrez?Db=pcsubstance&DbFrom=pcsubstance&Cmd=Link&LinkName=pcsubstance_pcsubstance_parent_popup&LinkReadableName=Same%20Parent&IdsFromResult=49854569&ordinalpos=1&itool=EntrezSystem2.PEntrez.Pcsubstance.Pcsubstance_ResultsPanel.Pcsubstance_RVDocSum’ “, “”, “”], [“Same Parent, Connectivity” , “window.top.location=’/sites/entrez?Db=pcsubstance&DbFrom=pcsubstance&Cmd=Link&LinkName=pcsubstance_pcsubstance_parent_connectivity_popup&LinkReadableName=Same%20Parent%2C%20Connectivity&IdsFromResult=49854569&ordinalpos=1&itool=EntrezSystem2.PEntrez.Pcsubstance.Pcsubstance_ResultsPanel.Pcsubstance_RVDocSum’ “, “”, “”], [“Similar Substances” , “window.top.location=’/sites/entrez?Db=pcsubstance&DbFrom=pcsubstance&Cmd=Link&LinkName=pcsubstance_pcsubstance&LinkReadableName=Similar%20Substances&IdsFromResult=49854569&ordinalpos=1&itool=EntrezSystem2.PEntrez.Pcsubstance.Pcsubstance_ResultsPanel.Pcsubstance_RVDocSum’ “, “”, “”], [“PubChem Same Compound” , “window.top.location=’/sites/entrez?Db=pccompound&DbFrom=pcsubstance&Cmd=Link&LinkName=pcsubstance_pccompound_same&LinkReadableName=PubChem%20Same%20Compound&IdsFromResult=49854569&ordinalpos=1&itool=EntrezSystem2.PEntrez.Pcsubstance.Pcsubstance_ResultsPanel.Pcsubstance_RVDocSum’ “, “”, “”], [“PubChem Component Compounds” , “window.top.location=’/sites/entrez?Db=pccompound&DbFrom=pcsubstance&Cmd=Link&LinkName=pcsubstance_pccompound&LinkReadableName=PubChem%20Component%20Compounds&IdsFromResult=49854569&ordinalpos=1&itool=EntrezSystem2.PEntrez.Pcsubstance.Pcsubstance_ResultsPanel.Pcsubstance_RVDocSum’ “, “”, “”] ] –>Related Structures, <!– var Menu49854569_4 = [ [“UseLocalConfig”, “jsmenu3Config”, “”, “”], [“PubMed MeSH Keyword Summary” , “window.top.location=’http://pubchem.ncbi.nlm.nih.gov/pmsummary/pubmed.cgi?db=pcsubstance&uid=49854569&itool=EntrezSystem2.PEntrez.Pcsubstance.Pcsubstance_ResultsPanel.Pcsubstance_RVDocSum&ordinalpos=1′ “, “”, “”], [“PMC Articles” , “window.top.location=’/sites/entrez?Db=pmc&DbFrom=pcsubstance&Cmd=Link&LinkName=pcsubstance_pmc&LinkReadableName=PMC%20Articles&IdsFromResult=49854569&ordinalpos=1&itool=EntrezSystem2.PEntrez.Pcsubstance.Pcsubstance_ResultsPanel.Pcsubstance_RVDocSum’ “, “”, “”], [“PubMed (MeSH Keyword)” , “window.top.location=’/sites/entrez?Db=pubmed&DbFrom=pcsubstance&Cmd=Link&LinkName=pcsubstance_pubmed_mesh&LinkReadableName=PubMed%20(MeSH%20Keyword)&IdsFromResult=49854569&ordinalpos=1&itool=EntrezSystem2.PEntrez.Pcsubstance.Pcsubstance_ResultsPanel.Pcsubstance_RVDocSum’ “, “”, “”], [“MeSH Keyword” , “window.top.location=’/sites/entrez?Db=mesh&DbFrom=pcsubstance&Cmd=Link&LinkName=pcsubstance_mesh&LinkReadableName=MeSH%20Keyword&IdsFromResult=49854569&ordinalpos=1&itool=EntrezSystem2.PEntrez.Pcsubstance.Pcsubstance_ResultsPanel.Pcsubstance_RVDocSum’ “, “”, “”] ] –>Literature<!– var PopUpMenu2_LocalConfig_jsmenu3Config = [ [“ShowCloseIcon”,”yes”], [“Help”,”window.open(‘/entrez/query/static/popup.html’,’Links_Help’,’resizable=no, scrollbars=yes, toolbar=no, location=no, directories=no, status=no, menubar=no, copyhistory=no, alwaysRaised=no, depend=no, width=400, height=500′);”], [“TitleText”,” Links “] ] var jsmenu3Config = [ [“UseLocalConfig”,”jsmenu3Config”,””,””] ] function ShowLinks(url,linkscount) { var X,Y; var H = (linkscount + 5)*30, W = 300; if(parseFloat(navigator.appVersion)>= 4) { if(navigator.appName==”Netscape”) { X=window.innerWidth;Y=window.innerHeight; if(H > window.innerHeight) { H=window.innerHeight-50;} }else{ X=document.body.offsetWidth;Y=document.body.offsetHeight; if(H > document.body.offsetHeight) { H=window.innerHeight-50;} } Y=(screen.height)/2-H/2; X=(screen.width)/2-W/2; } window.open(url, ‘Links’,’alwaysRaised=yes,screenX=’+String(X)+’,screenY=’+String(Y)+’,resizable=no,scrollbars=yes,toolbar=no,location=no,directories=no,status=no,menubar=no,title=no,copyhistory=yes,width=’+String(W)+’,height=’+String(H)).focus(); } –> |
||||
|
|||||
There is a lot of semantics we should encode here:
Pubchem ASSERTS that Leadscope (a depositor) has deposited a substance entry (SID: 49854569). Leadscope ASSERTS that this entry relates to the name “ferrocene”; that this entry relates to the name “catane” (and many more); that this entry is associated with formula C10H10Fe; that this entry has a connection table [identical with (c) above]; and so on.
The proper way to do this is with RDF using triples and/or reification, bnodes or quads. In this way we can see who asserted what or who asserted who said what. This is not chopping logic – there is no correct connection table for ferrocene , there are only assertions made by authorities (here Leadscope, although they may have taken it from somewhere else unspecified).
Let’s encode the formula. cml:formula is one of the benefits of CML and C4W has been written to manage formulae properly. There are 2 formulae, the sum of the atoms (concise) and an inline (which can be any text).
<molecule id="mol123456789" title="ferrocene" xmlns='http://www.xml-cml.org/schema'> <formula concise="C 10 H 10 Fe 1" inline="Fe(C_5_H_5)_2_"/> </molecule>
Now we come to the bonding. This representation has 3 species and CML supports sub-molecules (again an important feature). The iron is:
<molecule id="m1">
<atomArray>
<atom id="a0" elementType="Fe" formalCharge="2"/>
</atomArray>
</molecule>
That tells us everything about the iron with no implicit semantics. The Cps could be represented in a number of ways. I’ll write
<molecule id="m2" formalCharge="-1">
<atomArray>
<atom id="a1" elementType="C"/>
<atom id="a2" elementType="C"/>
<atom id="a3" elementType="C"/>
<atom id="a4" elementType="C"/>
<atom id="a5" elementType="C"/>
<atom id="a6" elementType="H"/>
<atom id="a7" elementType="H"/>
<atom id="a8" elementType="H"/>
<atom id="a9" elementType="H"/>
<atom id="a10" elementType="H"/>
</atomArray>
<bondArray>
<bond id="a1_a2" atomRefs2="a1 a2/>
<bond id="a2_a3" atomRefs2="a2 a3/>
<bond id="a3_a4" atomRefs2="a3 a4/>
<bond id="a4_a5" atomRefs2="a4 a5/>
<bond id="a5_a1" atomRefs2="a5 a1/>
<bond id="a1_a6" atomRefs2="a1 a6/>
<bond id="a2_a7" atomRefs2="a2 a7/>
<bond id="a3_a8" atomRefs2="a3 a8/>
<bond id="a4_a9" atomRefs2="a4 a9/>
<bond id="a5_a10" atomRefs2="a5 a10/>
</bondArray>
</molecule>
and the other by analogy. Note that I have not added any bond orders – this is deliberate. the community will argue about whether the ring bonds are single, double, delocalised, pi, etc. They will argue where the charge should be put. So I have added exactly enough information that stops before they start fighting.
Putting it together we get the complete CML for this particular representation:
<molecule id="mol123456789" title="ferrocene" xmlns='http://www.xml-cml.org/schema'>
<formula concise="C 10 H 10 Fe 1" inline="Fe(C_5_H_5)_2_"/>
<molecule id="m1">
<atomArray>
<atom id="a0" elementType="Fe" formalCharge="2"/>
</atomArray>
</molecule>
<molecule id="m2" formalCharge="-1">
<atomArray>
<atom id="a1" elementType="C"/>
<atom id="a2" elementType="C"/>
<atom id="a3" elementType="C"/>
<atom id="a4" elementType="C"/>
<atom id="a5" elementType="C"/>
<atom id="a6" elementType="H"/>
<atom id="a7" elementType="H"/>
<atom id="a8" elementType="H"/>
<atom id="a9" elementType="H"/>
<atom id="a10" elementType="H"/>
</atomArray>
<bondArray>
<bond id="a1_a2" atomRefs2="a1 a2"/>
<bond id="a2_a3" atomRefs2="a2 a3"/>
<bond id="a3_a4" atomRefs2="a3 a4"/>
<bond id="a4_a5" atomRefs2="a4 a5"/>
<bond id="a5_a1" atomRefs2="a5 a1"/>
<bond id="a1_a6" atomRefs2="a1 a6"/>
<bond id="a2_a7" atomRefs2="a2 a7"/>
<bond id="a3_a8" atomRefs2="a3 a8"/>
<bond id="a4_a9" atomRefs2="a4 a9"/>
<bond id="a5_a10" atomRefs2="a5 a10"/>
</bondArray>
</molecule>
<molecule id="m3" formalCharge="-1">
<atomArray>
<atom id="a11" elementType="C"/>
<atom id="a12" elementType="C"/>
<atom id="a13" elementType="C"/>
<atom id="a14" elementType="C"/>
<atom id="a15" elementType="C"/>
<atom id="a16" elementType="H"/>
<atom id="a17" elementType="H"/>
<atom id="a18" elementType="H"/>
<atom id="a19" elementType="H"/>
<atom id="a20" elementType="H"/>
</atomArray>
<bondArray>
<bond id="a11_a12" atomRefs2="a11 a12"/>
<bond id="a12_a13" atomRefs2="a12 a13"/>
<bond id="a13_a14" atomRefs2="a13 a14"/>
<bond id="a14_a15" atomRefs2="a14 a15"/>
<bond id="a15_a11" atomRefs2="a15 a11"/>
<bond id="a11_a16" atomRefs2="a11 a16"/>
<bond id="a12_a17" atomRefs2="a12 a17"/>
<bond id="a13_a18" atomRefs2="a13 a18"/>
<bond id="a14_a19" atomRefs2="a14 a19"/>
<bond id="a15_a20" atomRefs2="a15 a20"/>
</bondArray>
</molecule>
</molecule>
Notice how CML has represented exactly what we know about this depiction. (If I had wanted I could have put in the precise positions of the double bonds and carbanion, but it’s probably counterproductive). It’s completely semantic, no implicit information.
That’s enough for now; I promise that in the next post, Rich, I will deal with pi-bonding etc. I hope you will agree that this is one valid representation of ferrocene.