I’ve taken the chance pf a few days without commitments to investigate how we shall be using RDF. We’ve got several projects where we are starting to use it – CrystalEye – WWMM, eChemistry, SPECTRa : JISC and other ORE-based projects. I’ve been convinced for a few years that CML+RDF has to be the way forward for representing chemistry – the only question was when. CML gives the precision that is required for defining the local structure of objects (such as molecules) and RDF gives the flexibility for supporting a very diverse community who have different approaches and needs. It’s a balance between these two.
RDF represents information by triples – classically
subject – predicate – object
Here’s an example from WP:
<rdf:RDF xmlns:rdf="http://www.w3.org/1999/02/22-rdf-syntax-ns#" xmlns:dc="http://purl.org/dc/elements/1.1/"> <rdf:Description rdf:about="http://en.wikipedia.org/wiki/Tony_Benn"> <dc:title>Tony Benn</dc:title> <dc:publisher>Wikipedia</dc:publisher> </rdf:Description> </rdf:RDF>
To an English-speaking person, the same information could be represented simply as:
The title of this resource, which is published by Wikipedia, is ‘Tony Benn’
[Tony Benn is a well-known socialist UK politician much respected by people of all parties and none.]
This can be represented by a graph (from the W3C validator service) :

This is a very simple graph. The strength of RDF is that you can add a new triple anywhere and keep on doing it. The weakness of RDF is that you can add a new triple anywhere and keep on doing it. You end up with graphs of arbitrary structure. The challenge of ORE is to make sense of these.
Molecules have a variable RDF structure, We have to cater for molecules with no names, a hundred names, many properties, parameter constraints, etc. And the data are changing constantly and can come from many places. So there needs to be a versioning system and RDF is almost certainly the best way to tackle this. So here is a typical molecule:
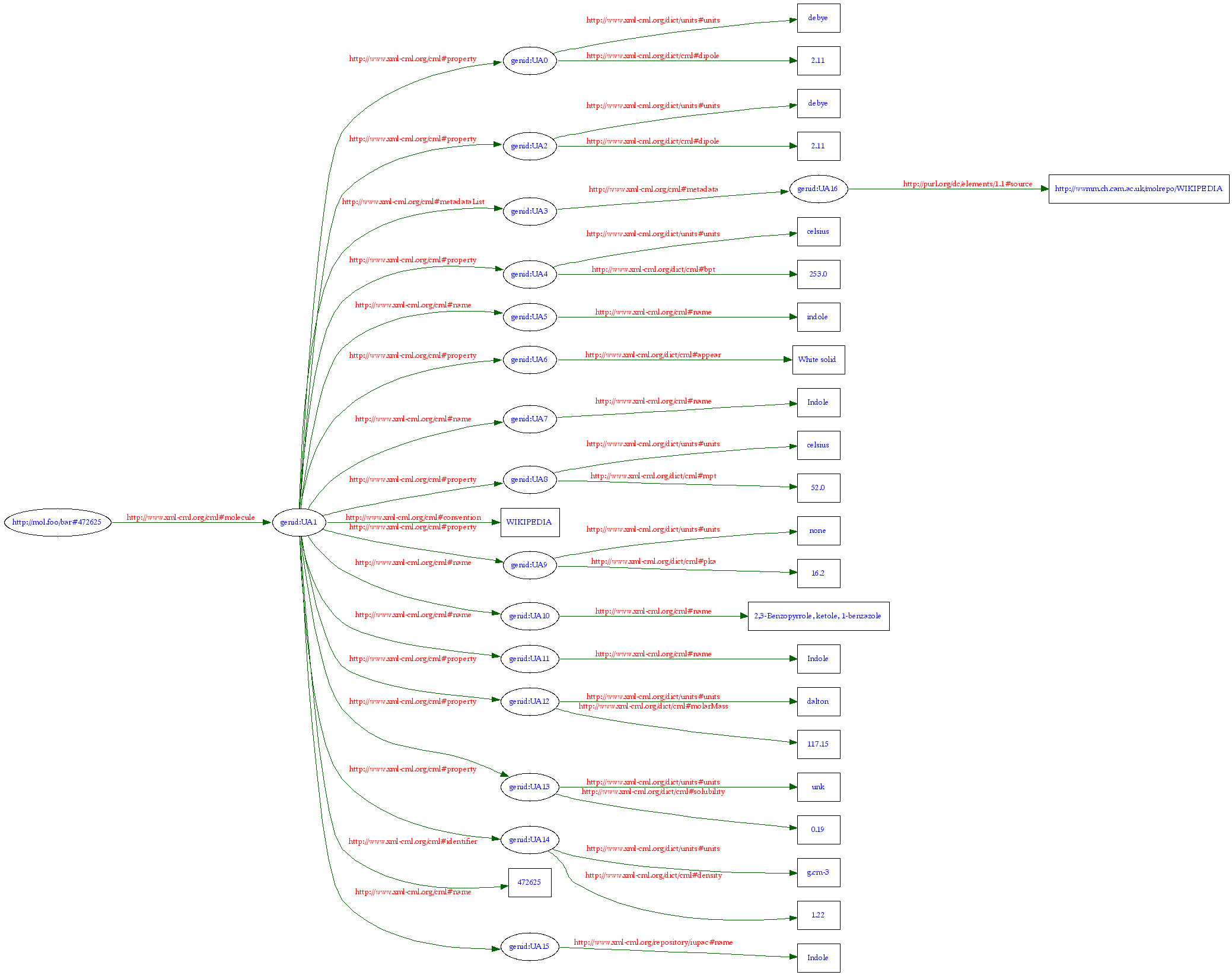
The quality is bad because the graph is much larger and had to be scaled down (you can click it). But it shows the general structure – a “molecule” node, with about 10 “properties” (in the RDF sense) and 3-4 layers.
The learning curve for RDF is steep. The nomenclature is abstract and takes some time to become familiar with. Irritatingly there are at least 4 different syntaxes and some parts of them are very similar. Several query languages as well. However having spent a day with Jena, I can now create RDF from CML and it makes a lot of sense. (Note that it’s relatively easy to create RDF from XML, but no guarantee that arbitrary RDF can be transformed to XML).
The key thing that you have to learn is that almost everything is a Uniform Resource Identifier (URI) or a literal. So up to now we have things in CML such as dictRef, convention, units. In RDF alll these have to be described by URIs. This is hard work but very good discipline and helps to firm up CML vocabulary and dictionaries.
So we now have over 100,000 chemical triples and should be able to do useful things very soon.
Is the molecule intended to have repeated instances of the same dipole?
(1) Thanks Joe, probably not 🙂
The information is derived from Wikipedia and even with the infoBox in XML parsing is not trivial. It is mots likely a bug in how I build the RDF graph.
(3) After some investigation it’s a feature in my software arising from the need to process several different forms of infobox. Since (at least at present) I don’t have specs for these I have had to guess what information comes where. Dipole can occur in different sections and the software has alternative approaches – both of these triggered in the current case. I’ve approached the chemists on WP and we should be able to clarify the formats.
Peter,
Sounds exciting!
Do you have any public RDF molecule data others could explore? URIs?
Tim BL
Pingback: Unilever Centre for Molecular Informatics, Cambridge - petermr’s blog » Blog Archive » Update on molecular repositories