I am delighted to announce and praise FROG, a free service for creating 3D molecular structures from connection tables. This is yet another module in the list of Open chemistry offerings that will ultimately give the tools to the chemistry community that they need and – possibly – deserve. I comment later.
var imagebase=’file://C:/Program Files/FeedReader30/’;
Recently, researchers at the French research institutes INSERM and CNRS developed an online service for converting SMILES string to 3D conformers: “FRee Online druG 3D conformation generator (Frog)”. A description of this service was published in T. Bohme Leite, D. Gomes, M.A. Miteva, J. Chomilier, B.O. Villoutreix and P. Tufféry. Nucleic Acids Research, 2007, 35, W568-W572:
Frog is an on-line service aimed at generating 3D conformations for drug-like compounds starting from their 1D or 2D descriptions. Given the atomic constitution of the molecules and connectivity information, Frog can identify the different unambiguous isomers corresponding to each compound, and generate single or multiple low-to-medium energy 3D conformations, using an assembly process that does not presently consider ring flexibility. Tests show that Frog is able to generate bioactive conformations close to those observed in crystallographic complexes.
On behalf of the OpenBabel project, I am pleased to announce that Dr. Bruno Villoutreix (INSERM, University of Paris 5) and Dr. Pierre Tufféry (INSERM, University of Paris 7) have generously donated their code to OpenBabel. This code will be incorporated into OpenBabel under the GPL in the coming months, making fast and accurate SMILES-to-3D conformer generation available to the open source community for the first time.
The absence of an open source 3D conformer generation algorithm has increasingly become a problem in recent years due to the popularity of SMILES strings for the description of molecular information. Fortunately, this problem has now been solved. Thanks again to all those involved in the development and release of this code.
For further information on Frog, please contact the corresponding author of the Frog paper.
PMR: FROG is exciting not just because of its functionality, but also because it’s OPEN SOURCE. It will be distributed under Open Babel, which uses the GPL license.
Why does this matter? Surely there have been free services like this before. What about PRODRG? (This is a service which has been going for at least 5 years, I think).
I make it clear that I have no quarrel with the authors and maintainers of PRODRG. However they ARE supported by Wellcome, who have blazed a trail for Open Access. Perhaps this is an opportunity to do the same for Open Source and Open Services.
Q: I’m in a non-academic (i.e. commercial) environment. Can I use this server for free?A: You are free to try a few test compounds (up to 5) – then request a license, as explained here
Q: I would like to use PRODRG locally, where can I download a copy?A: PRODRG executables are available under license, as explained here.
Q: I would like to run a database (> 50 compounds) of small molecules on your server. Is that OK?A: Please E-mail me first.
PMR: This shows all the worst aspects of “free” but not open services. You can try a few examples only (Wiley generously allowed me to inspect 3 spectra out of their collection of 500,000. Chemspider allows download of 100 molecules out of ca 10, 000, 000. The license is C20 – you have to fill in a form and then you get your personal copy of the software (presumably in binary – binary is a timebomb waiting to slef-destruct on the next OS upgrade). Can I convert 100 compounds? By default NO.
Maybe PRODRG should contact Wellcome and agree a method for making it Open. PRODRG was, of course, supported before the Wellcome insistence on Open Access, so maybe they can do something retrospectively.
Back to FROG. FROG Is Free. FROG is Open. If you don’t understand what this means, here is a simple translation.
The FROG code has been made available to the human race, without requiring payment now or in the future. ANYONE can have access to the source code. ANYONE can compile it, and ANYONE can run it. You don’t have to email the authors. You don’t have to request permission. You don’t have to tell anyone you are doing it. If you modify it you have to make your modifications available. You have to make clear what the original authors wrote and what you wrote. You have to give credit to the original authors. (There’s a bit more, but that’s the essence).
There is also a free service from FROG (“FRee Online druG 3D conformation generator (Frog)”.). Try it. Here’s penicillin (actually 6-aminopenicilanic acid):
C12C(N)C(=O)N1C(C(=O)O)C(C)(C)S2
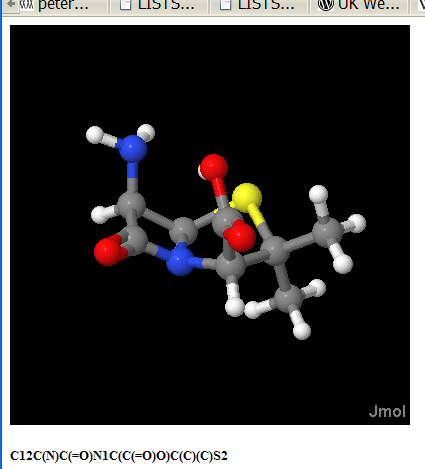
Who owns this image? YOU do. (Unless you donate it to a commercial publisher).
Are there restrictions on the FROG service? I don’t think so, but the service is completely separate from the code. Could they start charging in the future? Yes. Could they restrict access? Yes. Could they limit the usage? Yes. (I don’t think they will…)
Could I take the FROG code and run an Open service. Yes! Could I then close it and start charging money? Yes! Isn’t that immoral. No.
BECAUSE!!! We – the human race – now have the code. If something happens to the FROG service, we can duplicate it.
That’s why we are developing Open services at UCC. It’s not always trivial to distribute Open services (it’s harder than Open code). But we are thinking hard how to do it. Making the code Open is the major first step.

Please cease and desist from the spread of incorrect information about Chemspider.
You comment “Chemspider allows download of 100 molecules out of ca 10, 000, 000. ”
AS you commented in a later post “I do not follow Chemspider regularly”. Please clean this statement up in your multiple posts. The FAQ page at http://www.chemspider.com/FAQ.aspx declares
“May I download the data and use it in my own database(s)?
You have limited rights in this regard. You can only assemble a database of 5000 structures or less, and their associated properties, from our database without our permission. You can download up to 1000 structures per day from the website. Please contact us at feedbackATchemspiderDOTcom to request an extension outside this constraint. We are willing to provide the ENTIRE database of ChemSpider structures at your request – the file will consist of InChI Strings, InChIKeys and ChemSpider IDs. These constraints are under regular review so please feel free to engage us in conversation. ”
At this stage your comments are uninformed. the database is over 20 million structures at present but we ARE de-duplicating again after depositing a few million more structures.
You continue to ask people for comments on this blog. In this example you challenged Wiley http://wwmm.ch.cam.ac.uk/blogs/murrayrust/?p=554 but I have seen no response from them. Are you surprised? The vehemence in which you approach some of us is simply harsh. See Klaus’ comments here http://wwmm.ch.cam.ac.uk/blogs/murrayrust/?p=554#comment-45978. IF you have known the man for 20 years isn’t it more polite to call him on issues before publicly broadcasting disdain?
I have attempted to clean up your judgments regarding the fact that Wiley are deriving their data from the literature. I do not believe they are. You have not communicated this information to your audience. See http://www.chemspider.com/blog/?p=126
There are very few of us commenting on your blog posts. Your judgments are separating you from your audience. Peter..please take care. You have so much value to bring to us and so much good information to deliver please don’t taint it in this way.