I spent yesterday reviewing the data in BJOC (the Beilstein Journal of Organic Chemistry) (articles). This is a new (ca. 1 year) and important journal as it is the first free-to-author and free-to-read journal in chemistry, supported by the Beilstein Institut.
BJOC advocates Openness and has a facility for adding comments on each article. Up till now there has been little or no use of this facility (it is always daunting to be the first commentator) so I thought I would add some to catalyse the process of communal knowledge generation. I’m not an expert in most of the actual science, but I am familiar with the type of data in organic papers, which has a large emphasis on describing chemical reactions and the products.
.
.
.
.
.
.
[This space blank because the images are necessarily large and overlap the wordpress menu]
.
.
.
.
.
.
There is a fairly formal manner of reporting the chemical data:
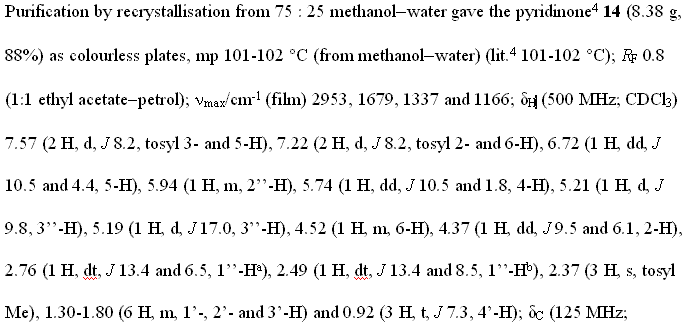
Not bedtime reading (except for chemists) but it is a formal statement of the experiment. (The tragedy is that it is used to be a machine-readable file with 16,000 points and that all chemical publishers require it to be abstracted into this text – but that is a different rant). The chemist has to extract this data from the spectrum, retype it into this form (taking ca. 45 minutes – my informal survey) and then sent to the editor. There can easily be 50 chunks of this stuff in one manuscript.
Because of this (absurb) retyping (instead of depositing the raw data with the publisher), errors creep in. The reviewer now has to check all this stuff by hand (again taking 30 minutes). And they really do – I have seen them. It could easily take a day to wade through the experimental section in a paper. Because this is the ultimate touchstone as to whether the correct compound has been made. (There have been frequent cases where claims of a synthesis have been later disproved by using this information – there was a recent example at the ACS meeting – entertainingly reviewed by Tenderbutton (search for “La Clair” in the text) where computer calculations on the proposed chemical structure did not agree with the published values). So they matter.
But what if they are mistyped? That’s where OSCAR comes in. Five years the Royal Society of Chemistry supported two undergraduate summer students (Joe Townsend and Fraser Norton) to look into creating an “Experimental Data Checker” for this sort of material. They did brilliantly and were followed next year by Sam Adams and Chris Waudby (also RSC funded). The result was OSCAR – the Open Source Chemical Analysis and Retrieval system, written in Java. Even if you are not a chemist, you may enjoy trying it out. You can take a raw manuscript (DOC) or published (HTML) – PDF is a hamburger so it probably won’t work – and drop it into OSCAR. OSCAR parses this text (using regular expressions) and produces:

OSCAR has recognised the different sort of spectra (coloured) the melting point, etc. It struggles with the appalling diversity of character sets used by Word (black squares) but makes sense of almost anything. These were very conscientious authors (they are from GSK) and the syntax is very correct. (This is impressive as there is no formal definition of the syntax, and OSCAR guesses from a large number of dialects).
OSCAR can now extract the data from the publication. Not, unfortunately, the raw data as the publishers currently don’t accept this for publication. But the numbers in the document. OSCAR can even guess what the spectrum might have looked like before it was lost in publication.
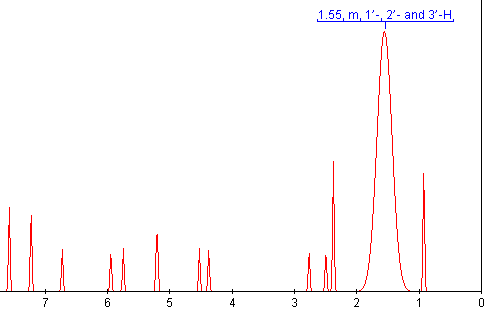
It’s only a guess, but hopefully it brings home what is lost in publication.
Now, although we don’t know whether any of the data are correct (only the author does) OSCAR has rules that point out when they might be incorrect. For example if a melting point of -300 deg C is reported it is obviously wrong (maybe a minus sign crept in). Or if the number of hydrogens calculated from the spectrum above don’t agree with the formula again something may be wrong. OSCAR applies these to each report of a chemical in the paper and lists all the warnings. Like CheckCIF (previous post) some of these are potential serious, while others are matters of style.
And, finally, OSCAR can extract all this data. I contend that the text above is factual material and cannot be copyrighted so OSCAR can extract data from the world’s chemical literature, even if it is closed and copyrighted. (However some publishers will probably claim that their articles are “entries in a database” and hence copyright. We have to fight this. It’s OSCAR that has done the work, not the publishers). The good news is (a) most data is now in open, non-copyright “Supplemental Material”, “Supporting Information”, or similar and (b) there are (a few) open access articles in chemistry. The next post shows how OSCAR can review the Open chemical literature…
(NB. This is OSCAR-1. The latest version, OSCAR3, has a much more sophisticated parser and can also work out chemical strucures from their names. It really is our vision of a “journal eating robot”. It’s on SourceForge but is strictly early adopter. More about that later.)
-
Recent Posts
-
Recent Comments
- pm286 on ContentMine at IFLA2017: The future of Libraries and Scholarly Communications
- Hiperterminal on ContentMine at IFLA2017: The future of Libraries and Scholarly Communications
- Next steps for Text & Data Mining | Unlocking Research on Text and Data Mining: Overview
- Publishers prioritize “self-plagiarism” detection over allowing new discoveries | Alex Holcombe's blog on Text and Data Mining: Overview
- Kytriya on Let’s get rid of CC-NC and CC-ND NOW! It really matters
-
Archives
- June 2018
- April 2018
- September 2017
- August 2017
- July 2017
- November 2016
- July 2016
- May 2016
- April 2016
- December 2015
- November 2015
- September 2015
- May 2015
- April 2015
- January 2015
- December 2014
- November 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- February 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- August 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- May 2007
- April 2007
- December 2006
- November 2006
- October 2006
- September 2006
-
Categories
- "virtual communities"
- ahm2007
- berlin5
- blueobelisk
- chemistry
- crystaleye
- cyberscience
- data
- etd2007
- fun
- general
- idcc3
- jisc-theorem
- mkm2007
- nmr
- open issues
- open notebook science
- oscar
- programming for scientists
- publishing
- puzzles
- repositories
- scifoo
- semanticWeb
- theses
- Uncategorized
- www2007
- XML
- xtech2007
-
Meta
Pingback: Unilever Centre for Molecular Informatics, Cambridge - petermr’s blog » Blog Archive » OSCAR reviews a journal
(1) This comment seems to have appeared because I referenced one of my own posts!
Yes – when you link to a post, WordPress (by default) tries to call out to it to see if it can send a trackback.
A good deal of the reasoning behind transcription of spectral data in publication is to impart meaning to the spectra. The 1H NMR spectrum of rasfonin, for instance, would be indeciferable to me, but the data written in the publication, transribed by the author and annoted for every peak would make (more) sense. It’s great to get an idea what the spectra look like, but more often than not, the actual spectra can be found in the supplementory data as a scan of the original. The combination of these two data sources gives the synthetic chemist everything they need.
(4) Thanks very much, totally synthetic. I agree completely that the annotation is critical. Part of my concern is that the annotation is not standardised and that spectra are not always deposited as supplemental. However the spectra are often deposited without axes or links to the annotation and are frequently difficult to interpret.
My main interest in doing this, however, is to preserve the spectra so that nyone can use them. It is technically possible to capture all the current spectra and to search and analyse them systematically. You, might, for example, have an unusual chemical shift and wish to see whether there is anything like it in the literature. In crystallography the systematic archiving of the data has led to over 1000 papers simply analysing the totality of the data. Spectra may not have as much information but I am sure that this would still be valuable.
P.
Pingback: Unilever Centre for Molecular Informatics, Cambridge - petermr’s blog » Blog Archive » Hamburger House of Horrors (1)