In my last post (Nature: How much content can our robots access?) I asked general questions about what data, if any, in a scientific article publishers would not allow humans and robots to use without permission. So, as an example, I’m asking Nature what parts of a data-rich article can be re-used automatically.
The model I have is that our robots can access electronic publications, extract data, and re-use it. Ideally we would like XML, but text and PDF can give good (though not perfect) results. Nature has recently brought out “Protocols”, which publishes recipes. I think this is a great idea – not enough credit is given to the actual work laboratory done in many subjects. So I give some snapshots (claiming fair use as the paper is freely readable and I haven’t taken the whole) show what our robots can do, and ask what I am permitted to do:
That’s the abstract. I expect it’s on Pubmed, so perhaps my robots can mine it for chemicals?
YES/NO?
This is one of the molecules mentioned in the paper (there are 10-20). This is the normal way of communicating the structure of a molecule, but it’s also an image. My robots can download it and interpret the structure. But technically it’s an image. Are the robots allowed to download and extract the molecular data without permission:
YES/NO
So then the protocol shows what actually happens in the reaction, and notes that there is a critical patch of colour:
Here the only way of communicating the FACT of the reaction colour is to show an image. This isn’t a creative work of art (though there are good photos and bad ones) but is an ESSENTIAL part of the record. There are now systems which could be trained to browse the Internet and return all flasks which contain a greenish liquid. Can we download and analyse images of experimental results:
YES/NO:
And here is the spectral data.
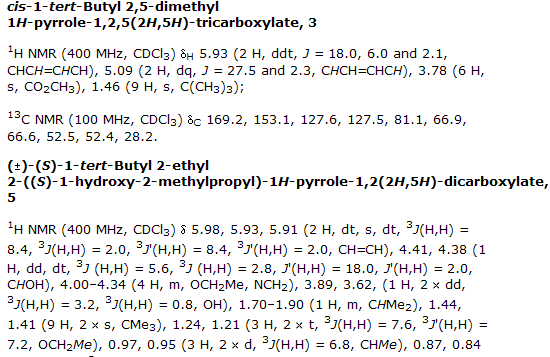
Can my robots download this and turn it into semantic data.?
And in doing so this is the sort of thing that OSCAR can produce. It’s taken part of the text and identified all the compounds (and worked out the atomic connectivity – see “furan” at the right).
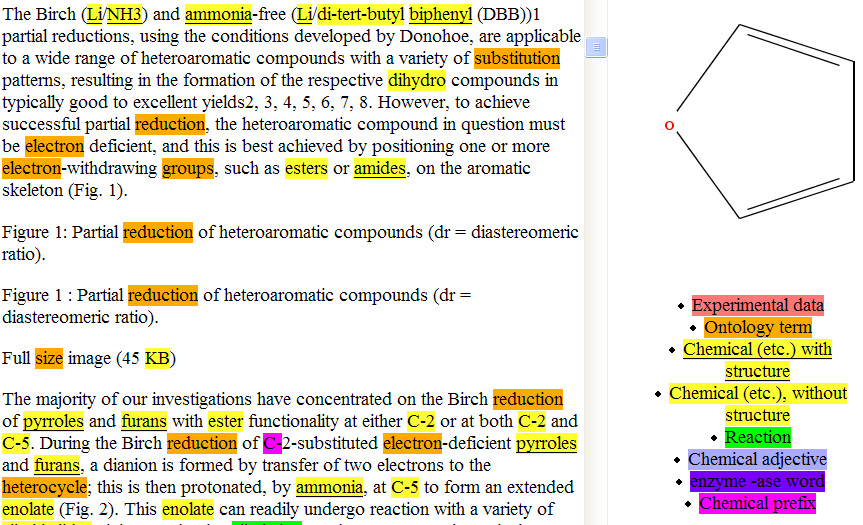
All of this is enormously valuable. If we are allowed to do it we can answer questions like “what reactions involve furans, or pyrroles, use Sodium ammonia, and undergo a light green colour”.
But it will only happen if publishers let us have access to their data without any barrier – no written permissions, no hangup over scientific photographs and images.
I’ve put Timo on the spot, but I trust him to be clear. That will make it easier when asking the other publishers later.
-
Recent Posts
-
Recent Comments
- pm286 on ContentMine at IFLA2017: The future of Libraries and Scholarly Communications
- Hiperterminal on ContentMine at IFLA2017: The future of Libraries and Scholarly Communications
- Next steps for Text & Data Mining | Unlocking Research on Text and Data Mining: Overview
- Publishers prioritize “self-plagiarism” detection over allowing new discoveries | Alex Holcombe's blog on Text and Data Mining: Overview
- Kytriya on Let’s get rid of CC-NC and CC-ND NOW! It really matters
-
Archives
- June 2018
- April 2018
- September 2017
- August 2017
- July 2017
- November 2016
- July 2016
- May 2016
- April 2016
- December 2015
- November 2015
- September 2015
- May 2015
- April 2015
- January 2015
- December 2014
- November 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- February 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- August 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- May 2007
- April 2007
- December 2006
- November 2006
- October 2006
- September 2006
-
Categories
- "virtual communities"
- ahm2007
- berlin5
- blueobelisk
- chemistry
- crystaleye
- cyberscience
- data
- etd2007
- fun
- general
- idcc3
- jisc-theorem
- mkm2007
- nmr
- open issues
- open notebook science
- oscar
- programming for scientists
- publishing
- puzzles
- repositories
- scifoo
- semanticWeb
- theses
- Uncategorized
- www2007
- XML
- xtech2007
-
Meta