It’s often said by detractors and obfuscates that “there is no demand for content mining”. It’s difficult to show demand for something that isn’t widely available and which people have been scared to use publicly. So this is an occasional post to show the very varied things that content mining can do.
It wouldn’t be difficult to make a list of 101 things that a book can be used for. Or television. Or a computer (remember when IBM told the world that it only needed 10 computers?) Content mining of the public Internet is no different.
I’m listing them in the order they come into my head, and varying them. The primary target will be scientific publications (open or closed – FACTs cannot be copyrighted) but the technology can be applied to government documents, catalogues, newspapers, etc. Since most people probably limit “content” to words in the text (e.g. in a search engine) I’ll try to enlarge the vision. I’ll put in brackets the scale of the problem
- Which universities in SE Asia do scientists from Cambridge work with? (We get asked this sort of thing regularly by ViceChancellors). By examining the list of authors of papers from Cambridge and the affiliations of their co-authors we can get a very good approximation. (Feasible now).
-
Which papers contain grayscale images which could be interpreted as Gels? A http://en.wikipedia.org/wiki/Polyacrylamide_gel is a universal method of identifying proteins and other biomolecules. A typical gel (Wikipedia CC-BY-SA) looks like
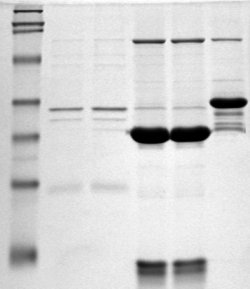 Literally millions of such gels are published each year and they are highly diagnostic for molecular biology. They are always grayscale and have vertical tracks, so very characteristic. (Feasibility – good summer student project in simple computer vision using histograms).
Literally millions of such gels are published each year and they are highly diagnostic for molecular biology. They are always grayscale and have vertical tracks, so very characteristic. (Feasibility – good summer student project in simple computer vision using histograms). - Find me papers in subjects which are (not) editorials, news, corrections, retractions, reviews, etc. Slightly journal/publisher-dependent but otherwise very simple.
- Find papers about chemistry in the German language. Highly tractable. Typical approach would be to find the 50 commonest words (e.g. “ein”, “das”,…) in a paper and show the frequency is very different from English (“one”, “the” …)
- Find references to papers by a given author. This is metadata and therefore FACTual. It is usually trivial to extract references and authors. More difficult, of course to disambiguate.
- Find uses of the term “Open Data” before 2006. Remarkably the term was almost unknown before 2006 when I started a Wikipedia article on it.
- Find papers where authors come from chemistry department(s) and a linguistics department. Easyish (assuming the departments have reasonable names and you have some aliases (“Molecular Sciences”, “Biochemistry”)…)
- Find papers acknowledging support from the Wellcome Trust. (So we can check for OA compliance…).
- Find papers with supplemental data files. Journal-specific but easily scalable.
- Find papers with embedded mathematics. Lots of possible approaches. Equations are often whitespaced, text contains non-ASCII characters (e.g. greeks, scripts, aleph, etc.) Heavy use of sub- and superscripts. A fun project for an enthusiast
So that’s just a start. I can probably get to 50 fairly easily but I’d love to have ideas from…
…YOU
[The title many or may not allude to http://en.wikipedia.org/wiki/101_Uses_for_a_Dead_Cat ]
Would it be very difficult to automaticaly detect graphs in papers and extract the values of the data points? I am now extracting (by hand) those data from an Arhenius plot, which I intend to use for an Eyring plot 😉
That is exactly what we plan.
It depends on the graph.
(a) is it vector (in PDF) or pixel PNG/JPG
(b) is all the detail visible?
(c) how much overlap
Can you send an example? Ideally one which is visible on the web.
There are so many graph variations that this would be hard. Just consider the problem of graphs with different shaped points – identifying them and then finding their centers. IMO, this is better done as an app that loads the graph image and key points are then extracted by hand and the values calculated by the code. My sense is that this would make a great crowd project – lots of individuals doing the work for each chart.
We can do it automatically for simple non-crowded x-y graphs. We’ll see how far this takes us
You might want to take a random sampling of charts to see how feasible this is. I looked at it about a decade ago when I co-founded a data start-up. Maybe I wasn’t inventive enough, but it looked like the only viable solution to me was an application assisted human to speed the process.
See graph 3.c from dx.doi.org/10.1074/jbc.M708010200
I opened the PDF, zoomed as much as possible, took a “print screen” and then measured the data position “by hand”.
That’s actually a scalable diagram in the PDF so will be arty straightforward to extract. Perhaps 2-4 weeks from now till the software is available
That figure is also available as part of a larger JPG in the HTML version of the paper.
Other uses for content mining:
– finding papers with EPR spectra and extracting each spectrum for properly named files.
– Find cartesian coordinates of molecules in Supporting Information PDF files and convert all of them them to a single file in the popular XYZ format so that all geometries may be easily analyzed with molecular visualization software.
– Track nomenclatural changes of microorganisms across the literature, build a central “synonymy list” so that any search for (e.g.) Cupriavidus metallidurans automatically retrieves the literature for its synonyms Ralstonia eutropha, Ralstonia metallidurans and Alcaligenes eutrophus.
– collating the results of all redox titrations of metallic sites in proteins, classified by protein, metal center, technique (EPR-monitored, UV-Vis, electrochemistry) and source.
Great ideas,
Feasibility depends slightly on whether info is in full text or suppinfo. The former is more likely to be vectors but more variable.
– finding papers with EPR spectra and extracting each spectrum for properly named files.
If vectors we should get the full set of data to same resolution as published. If pixels it will be worse than 1 pixel.
– Find cartesian coordinates of molecules in Supporting Information PDF files and convert all of them them to a single file in the popular XYZ format so that all geometries may be easily analyzed with molecular visualization software.
That will be very valuable and we can also archive them in Crystallography Open Database. We are starting to take theoretical coords there.
– Track nomenclatural changes of microorganisms across the literature, build a central “synonymy list” so that any search for (e.g.) Cupriavidus metallidurans automatically retrieves the literature for its synonyms Ralstonia eutropha, Ralstonia metallidurans and Alcaligenes eutrophus.
We can extract names with high precision/recall but will have to leave reconciliation to experts.
– collating the results of all redox titrations of metallic sites in proteins, classified by protein, metal center, technique (EPR-monitored, UV-Vis, electrochemistry) and source.
I would need a small corpus to comment on. The greater the consistency of presentation the easier.
“- collating the results of all redox titrations of metallic sites in proteins, classified by protein, metal center, technique (EPR-monitored, UV-Vis, electrochemistry) and source.
I would need a small corpus to comment on. The greater the consistency of presentation the easier.”
Here you have a relatively varied corpus:
10.1021/bi4009903
10.1021/bi4009903
10.1111/j.1432-1033.1997.00454.x
10.1021/bi401305w
10.1093/jb/mvt026
10.1021/bi00217a037
10.1021/bi00127a015
I believe that a full tezt search for the words “mV” or “vs. standard hydrogen electrode” in the vicinity of “redox titration” would be enough to collate a very complete table of data …
This is very useful. We can certainly use the phrases to retrieve the paper. It may be possible to extract actual data. It may be possible to link it within tables. Whether it’s possible to pull out more structured info – like the sites in the protein I don’t know.
You have probably already thought about this: extracting the specifics of computational chemistry calculations, which are typically set in prose.
Yes.
I really we would like to use the log files where we can extract massive amounts of data. But it’s a question of how formulaic the language and tables are.
Pingback: Four short links: 26 May 2014 - O'Reilly Radar
I don’t think the protein gel recognition even warrants a summer project. The images are quite distinctive – horizontal bands with similar pixel colors across much of the x axis. The markers should be easy to detect within the image. The PLOS site would be a good resource to test the methodology and code.
I agree it’s relatively easy to recognise these from intensity and spatial frequencies but the parameters will have to be set and hopefully by adaptive methods. That needs a student or equivalent
I have done this before but with different data – P33 macroarray gene expression data. It isn’t that hard, even with spots rather than bands, and that included quantification (spot size and density)
2D protein gel identification would be another interesting target. That might well be a bigger project.
So what sort of queries are you expecting with gel electrophoresis identification? Find me all papers with mention of protein X AND with a protein gel image as part of the content?
Simplest to most difficult:
* is this figure a gel? Use captions and/or images
* how many lanes are there and what are they labelled (needs caption)
* how many bands are there in each lane? Extract density of each
* has this gel been photoshopped (fraud)
One issue that occurs to me is that protein gel images are likely to be confused with northern blot (amongst others) images. Does that matter, or would gel electrophoresis images be the recognition target?
I think you have answered my question. You want the caption to be read too, to identify the image and details of the result.
See what Ross Mounce has done at https://www.flickr.com/photos/123636286@N02/sets/ – all the caption metadata is embedded in the image – we are interpreting the figures as (binary) trees which may have explicit species or may be labelled by captions
Don’t know offhand. We are going to use caption information as well as the image which should increase the specificity. And unless the gel is labelled to some extent then it only acts as an indicator of the theme of the paper. “This paper contains gels and crystal structures” so it’s probably structural biology
So as long as the caption is text in the body, identified with a Figure number, he can capture it. That seems reasonably straightforward, although positioning of teh caption may be an issue. A number of images have no source or caption (e.g. https://www.flickr.com/photos/123636286@N02/14319160853/ ). I am not clear why neither the source nor the caption was captured.
Still, nice work.
In the era when the cloud was still nascent, I knew a company that was building a database to relate text and images for rapid search. I don’t know what happened to them (and I forget the name) but I hope they were successful and migrated to the cloud, as the possibilities were cool and meshed with the web based chemogenomics database the company I worked for had built. Where we relied purely on created numeric and text data, this other company could add images of 2D gels, x-rays, charts, etc., necessary for inspection before believing the data.
Turns out that (unknown to the publisher) the caption is embedded in the image. Obviously this one escaped. But we are also capturing by hard work as well
I didn’t realize images submitted for publication could/would have caption metadata attached. Interesting. I assumed that they would be simple file formats – jpg/png/bmp etc and that any information about the image would be added to the text body or be part of the image (e.g. annotation).
Are you putting together something in this regard?