Typed in support of a community that cares passionately into Arcturus
Here’s an excellent example of a community that is racing towards mandating data deposition as part of publication. And to give credit where it’s due it’s reported by an ACS journal (J. Proteome Research) http://pubs.acs.org/doi/full/10.1021/pr900912g which is also slowly picking up the momentum.
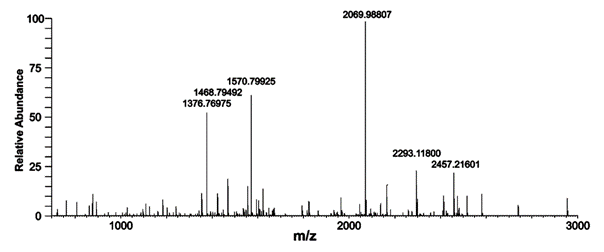
Don’t switch off at “Proteome”. The discipline is about finding what proteins are produced in living organisms and often how they affect disease or act as signals for it. The subject is data-rich as it involves injecting samples into a mass spectrometer; from the peaks in the spectrum (see image, stay with me) it’s possible to identify the proteins. So the data really matter.
- Is the experiment valid? A less-than-careful scientist could get impurities in the sample (or other artefacts) and come to the wrong conclusion. The data are a major step in allowing the referees and later readers to decide whether the experiment might be flawed.
- There is huge potential for re-using the data. This could be between species, cell types, diseases, etc. It’s still at an early stage because the data are complex but I am certain it will happen.
I quote selected chunks from the article (pleading fair use, especially as the article is Freely (if not Openly) published) . Here’s a spectrum – a piece of FACTUAL data (not a creative work) lifted without permission or shame or foreboding from the ACS. The peaks in the spectrum are chopped up proteins (peptides) from which the proteins can be identified. I make a few comments at the end
 ely (if not Openly) published).lished). (pleading fair use, especially as the o-auth eases, etc. It’ong conclusion. The data a
ely (if not Openly) published).lished). (pleading fair use, especially as the o-auth eases, etc. It’ong conclusion. The data a
MCP ups the ante by mandating raw-data deposition
J. Proteome Res., 2009, 8 (11), pp 4887–4888 DOI: 10.1021/pr900912g Copyright © 2009 American Chemical Society
Yet again, the editors of Molecular & Cellular Proteomics (MCP) are leading the way toward stricter publishing requirements for the proteomics field. At the First International Forum of Proteomics on September 26, 2009, and then two days later at the HUPO Eighth Annual World Congress (both held in Toronto), MCP Co-Editor Ralph A. Bradshaw announced that the journal will require its authors to deposit supporting raw MS data into a public repository at the time of publication. The new mandate will likely take effect in January 2010.
[…]
The ultimate goal of the mandate for raw-data deposition is to provide further proof that proteomics is a valid pursuit. “There’s an important sense that we need this in order to make proteomics more legitimate than it presently is,” says Bradshaw. “We think we need to raise the bar even further in terms of getting the proteomics literature validated and getting rid of the errors.”
[…] An increase in raw-data submissions would be a boon for bioinformaticians, who are always hungry for more data sets for tool development and testing. Philip Andrews of the University of Michigan adds that some valid identifications might be salvaged if researchers could reevaluate data from published experiments with other algorithms. In addition, some useful information, such as data about the instrument’s parameters, is lost when data are processed.
[…]Daniel Figeys of the Ottawa Institute of Systems Biology says that, in theory, raw-data deposition shouldn’t be a big deal for proteomics researchers. After all, researchers in the gene-expression field have been required to deposit all of their data into public databases for years. Also, the infrastructure for handling this type of proteomics data has been evolving and improving, says Andrews, who is the principal investigator of the Tranche Project. (Tranche is a repository for the long-term storage of data of any format.)
… The Amsterdam Principles, […] encourage open access to proteomic data. (Disclosure: K. Cottingham is a o-author of this manuscript.) …. “The community is going from asking questions such as, ‘Should there be data-release policies for proteomics?’ to asking, ‘What are the policies, and how and when will they finally be implemented?’ ” he explains.
” he explains.
… researchers have uploaded 10.8 terabytes of proteomics data into Tranche. Researchers also are using the data. “Since February,  8 terabytes of data have been downloaded by investigators, and that’s a lot of data to be downloaded from a repository like this,” he explains.
8 terabytes of data have been downloaded by investigators, and that’s a lot of data to be downloaded from a repository like this,” he explains.
Although Bradshaw says that MCP will not specify that researchers must upload the raw data to Tranche, he admits that it is “probably the only place where this would work” currently; other proteomics repositories and databases accept only processed data. Because Tranche is part of the ProteomExchange consortium, however, other resources, such as PRIDE and Peptidome, have access to its data and routinely download new data sets.
“I think the biggest concern about Tranche is that it’s supported with soft money, and it doesn’t have
Bradshaw hopes that researchers won’t view the requirement as a hardship and says, “The last thing in the world we want to do is create burdens and obstacles for authors.” Andrews explains that researchers uploading data to Tranche for the first time may experience a learning curve, but “once they realize how easy it is, I don’t think they’re going to be too concerned.”
-
Top of Page
-
Why make this move?
-
Data in, data out
-
Will other journals at the table follow suit?
-
MCP’s hand: on the table for public comment
…Proteomics and Proteomics: Clinical Applications strongly encourage the submission of processed data, but change is coming. Michael Dunn, who is at University College Dublin and is editor-in-chief of these journals, says, “We now propose that we will make a change in our next release of the Instructions to Authors, for January 2010, to state that we strongly encourage deposition of raw and processed data…
JPR‘s editor-in-chief, William Hancock … “My position was that we would strongly encourage authors and reviewers to support this initiative.”
-
Top of Page
-
Why make this move?
-
Data in, data out
-
Will other journals at the table follow suit?
-
MCP’s hand: on the table for public comment
Right now, all of the proposed changes exist in draft form and will be posted soon on the MCP website for public comment. Bradshaw explains, “Some of this could change, but I feel very confident in saying that the mandatory [raw-data] requirement is not going to change.”
We know Ralph very well and I have close relations with a scientist who reviews for MCP. When we visited Ralph earlier this year I told him about the Panton Principles (http://pantonprinciples.org/ ) and he enthusiastically communicated them to Tranche.
a big problem with this effort is the size of the datasets. I routinely generate RAW data for an experiment that is in the hundreds of gigabytes. I’ve tried some of the public databases and none of them can support data-transfer speeds that are sufficient for transfers that large. the other problem is proprietary data-formats for RAW data. i’m sure the field will get there, but at the moment it’s not JUST investigators that are protective of their data… there are significant technical hurdles as well.